Extended Data Fig. 7 |. Co-expression network module replication and tissue associations with CAD traits.
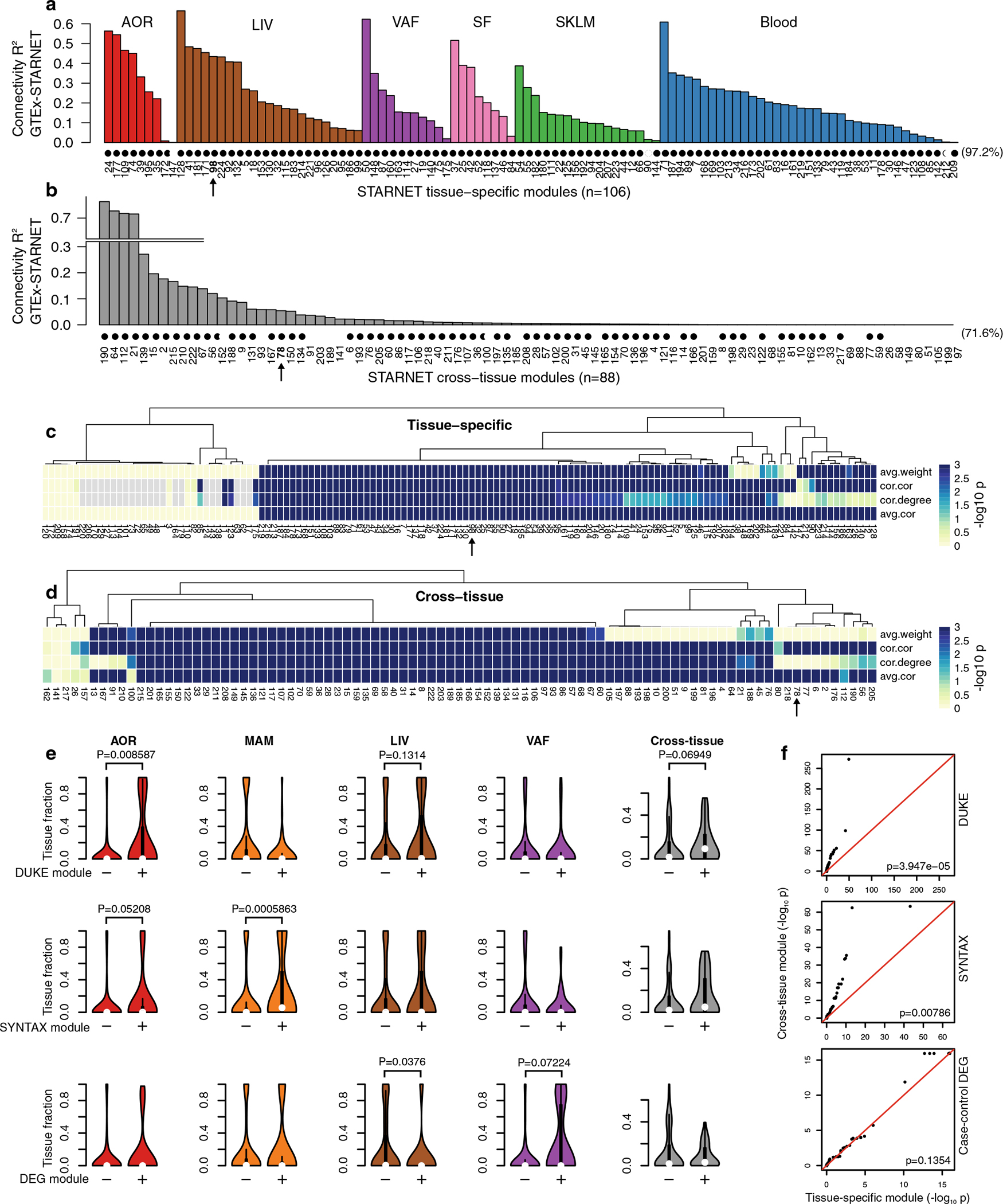
a, Concordance with GTEx RNA-seq data of gene-gene correlation coefficients in tissue-specific modules and in, b, cross-tissue modules, excluding gene-gene interactions within tissue. Modules indicated with black circle showed significant similarity at FDR < 0.05, Bonferroni corrected for n = 224 tests. c, d, In the absence of normal distributions of network correlation coefficients and weights, we also used 4 one-sided permutation tests implemented in the NetRep R package21 for replication of tissue-specific (c) and cross-tissue (d) co-expression modules. The scale is −log10 P values from the permutation test for the 4 different measures of network validation, where the extreme corresponds to P < 0.001 indicating the maximum sensitivity based on 1,000 permutations. e, Violin plots comparing tissue fractions of genes in co-expression modules that are significantly associated (+) with SYNTAX score (upper panel), Duke score (middle panel), or enriched (+) in differentially expressed CAD genes (Hypergeometric test) (Fig. 2c) with the tissue fraction of module genes that are not (−) (FDR < 0.05). P-values comparing the ‘+’and “−”module groups are calculated using Wilcoxon two-sided signed-rank test. For SYNTAX and Duke scores, which estimate arterial atherosclerosis, the co-expression modules were associated by aggregating gene-level P-values using Fisher’s method. f, QQ plots comparing tissue-specific and cross-tissue co-expression module aggregated P-values (Fisher’s method) for Duke (upper panel) and SYNTAX score (middle panel) and module enrichment of CAD DEGs. In contrast to CAD DEGs, cross-tissue modules are significantly more associated with Duke and SYNTAX scores than tissue-specific modules (Wilcoxon rank-sum test).
