Fig. 5 |. Endocrine factors in cross-tissue networks mediate signaling between GRNs in metabolic tissues and the arterial wall.
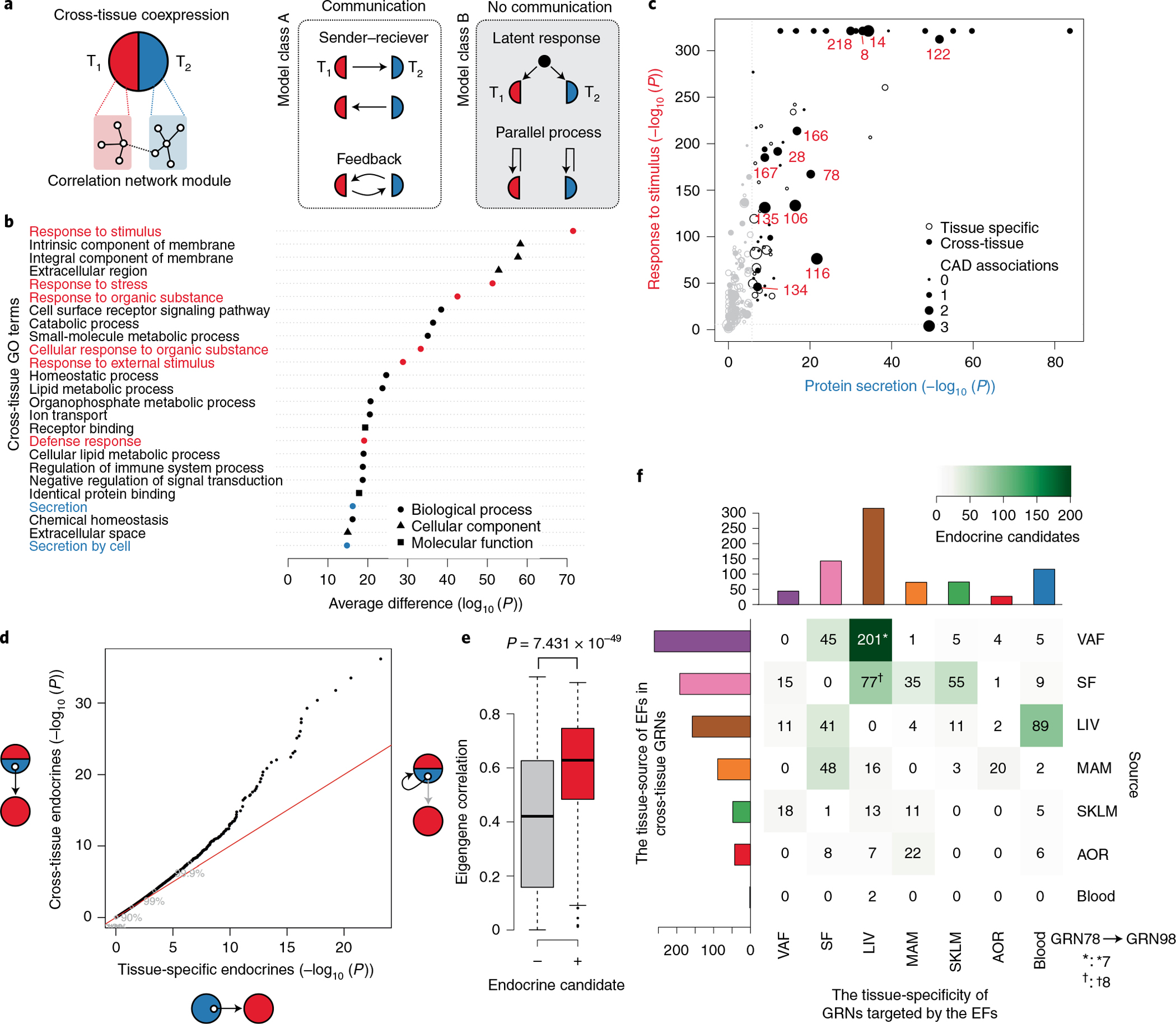
a, Two biological models that may explain cross-tissue GRNs. Left, communicative models, such as sender–receiver or feedback signaling. Right, noncommunicative models, such as parallel coincidental responses to latent external factors (for example, circulating or neuronal factors) or parallel but independent intra-tissue processes such as cell mitosis. T1, tissue 1; T2, tissue 2. b, Difference in GO enrichment between cross-tissue and tissue-specific GRNs (Fisher’s exact test, one-sided Mann–Whitney test of enrichment log (P values), FDR < 0.01). Enrichments of GO terms related to cellular signal reception and transmission are marked in red and blue, respectively. c, GO enrichments of cross-tissue and tissue-specific GRNs for the sender term ‘protein secretion’ (x axis) and the receiver term ‘response to stimuli’ (y axis). CAD-associated GRNs are marked in red. Gene set enrichment was performed by Fisher’s exact test. d, Quantile–quantile plot of correlations between genes (mRNA) encoding secreted proteins (UniProt annotation) in cross-tissue (y axis) and tissue-specific (x axis) GRNs and eigengene values of tissue-specific GRNs separate from the origin of the mRNA of the secreted protein (Pearson correlations, two-tailed t-test). e, Box plot showing average mRNA correlations of genes encoding secreted proteins with the eigengene of their host cross-tissue GRNs, comparing secreted proteins defined as ‘endocrine candidates’ (+, n = 467) and those that were not (−, n = 2,959). ‘Endocrine candidates’ were defined as secreted proteins in cross-tissue GRNs with correlations with tissue-specific GRNs at FDR < 0.2 in d. Median, center; lower and upper quartiles, box; 1.5 × interquartile range, whiskers. Two-sample Wilcoxon test. f, Heatmap of a tissue source–target matrix of 793 significant associations between pairs of endocrine candidate mRNA expression values and eigengene values of tissue-specific GRN targets. In total, 374 unique endocrine candidate genes were identified. 13 of these endocrine candidates identified in VAF (n = 7†) or SF (n = 8*) as part of the cross-tissue (liver-SF-VAF) GRN78 targeted the liver-specific GRN98 representing the most reliable edge in the supernetwork according to the bootstrapping analysis (Extended Data Fig. 10h).
