Extended Data Figure 7: Mutation frequency and percentage of hypermutated E/SEs in BL and CLL.
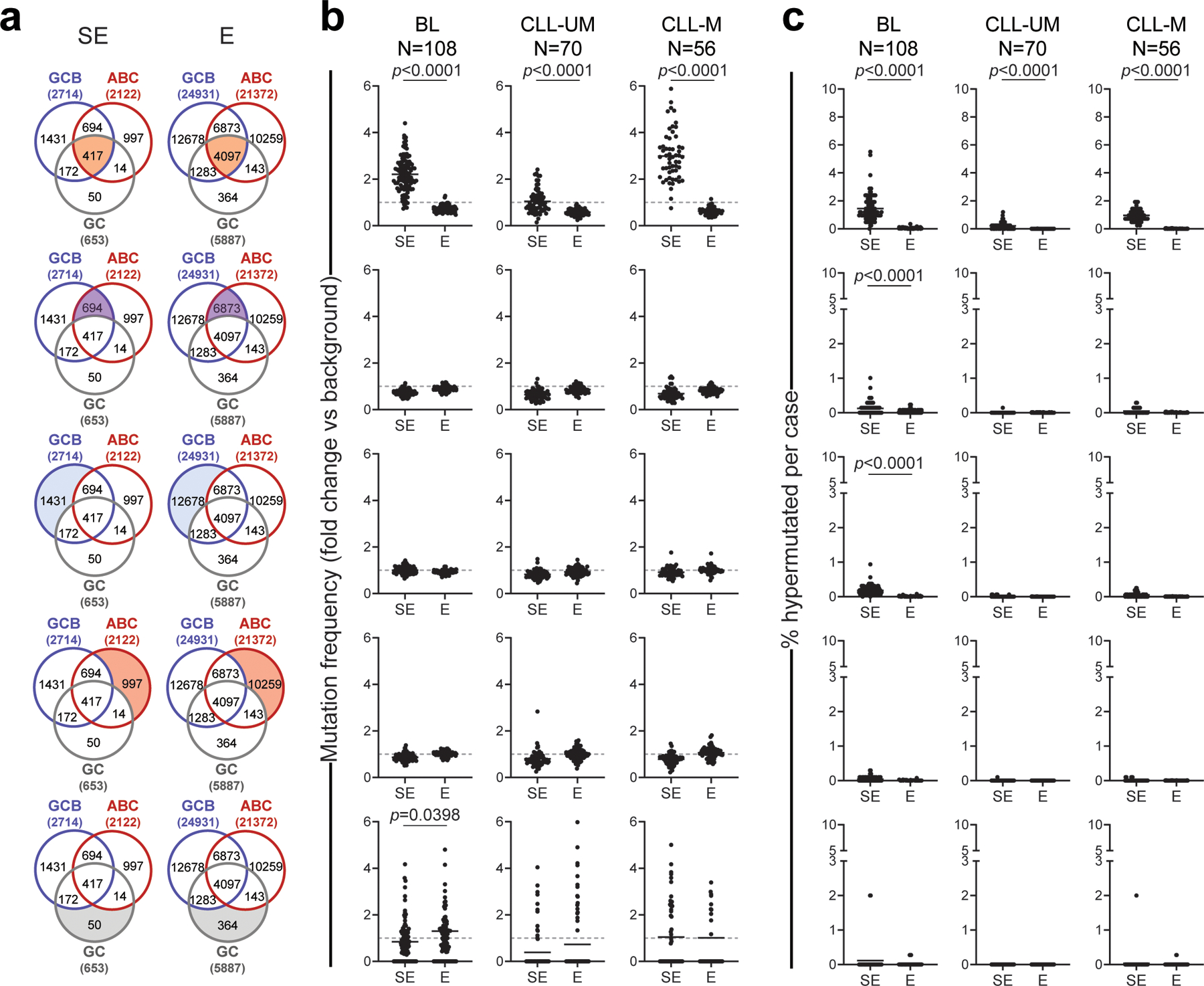
a. Venn diagrams showing the overlap between E/SEs identified in normal GC B cells, GCB-DLBCL cell lines and ABC-DLBCL cell lines. The shadowed area marks the subset of E/SEs interrogated in each of the analyses shown in panels b and c (same row), and the corresponding number is given for each subset inside the diagram. The total number of E/SEs identified in GC B cells, GCB-DLBCL cell lines and ABC-DLBCL cell lines appears outside the Venn diagram, in brackets. b, c. Sample-based mutation frequency (b) and percentage (c) of hypermutated E/SEs in primary BL and CLL specimens grouped based on molecular subtypes (IGHV-unmutated: CLL-UM; IGHV-mutated: CLL-M). The analysis of different regions (corresponding to those highlighted in the aligned Venn diagram) is displayed in different rows. In the graphs, each dot denotes one primary biopsy, and mutation frequencies are expressed as fold changes vs. background, calculated in the same sample as described in Methods. The grey dashed line in panel b represents the background mutation frequency, set at 1 for each sample (see Methods). All p-values were calculated by two-sided Wilcoxon rank-sum test after BH correction. Note that 82% of hypermutated SEs in CLL map to the IG loci.
