Figure 6: A mutational hotspot in the CXCR4 SE disrupts NR3C1 binding and transcriptional repression.
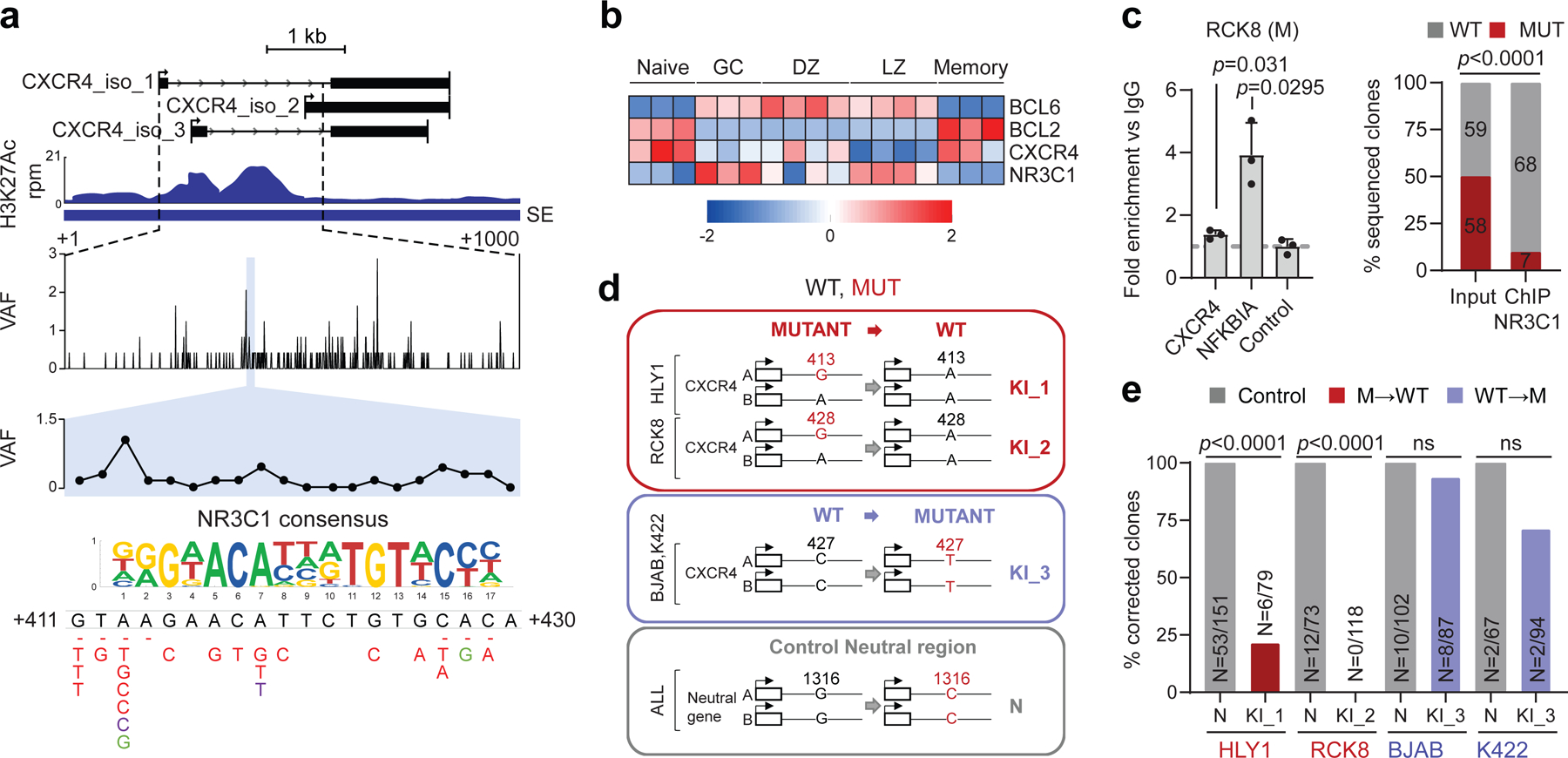
H3K27Ac ChIP-seq track of the CXCR4 iSE in LY10 (top). The hypermutated region is magnified below the track to show the VAF in primary DLBCL cases, and the mutational hotspot corresponding to the predicted NR3C1 consensus binding motif is expanded further (blue shadow; position according to NM_003467). SNVs found in DLBCL are positioned below (red, WGS data; green, cell lines; dotted line, deletion). b. Relative expression of BCL6, BCL2, CXCR4, and NR3C1 in normal B-cell subsets (z-scored log2 TPM). c. NR3C1 ChIP-qPCR (left) and allelic quantification of input and NR3C1-IP DNA (right) in the mutant RCK8 cell line, assessed by PCR amplification and cloning (one representative experiment in triplicate, out of two that gave similar results). d. Design of the CRISPR-Cas9 experiment utilized to correct the CXCR4 hotspot mutations in RCK8 and HLY1. e. Normalized percentage of corrected clones recovered in the CRISPR experiment; the percentage of properly mutated clones in the neutral region is set as 100% and the absolute clone numbers are indicated inside the bars. P-values were calculated by one-way ANOVA with Bonferroni correction (c, right panel), and two-tailed Fisher’s exact test (c, left panel, and e).
