Figure 1: Identification of functional NT sources to the BLA in associative learning.
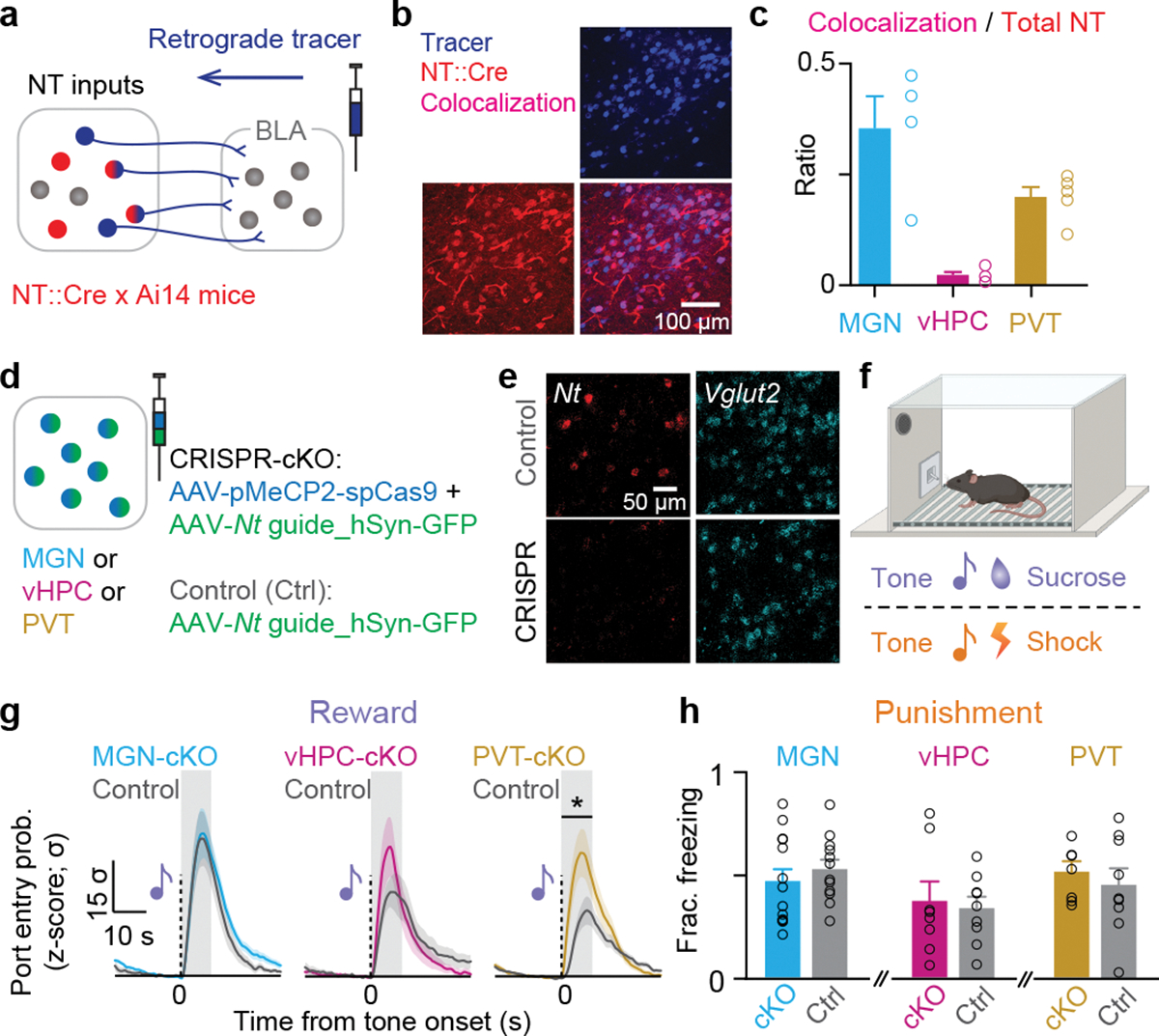
(a) Schematic of viral injection. (b) Representative confocal image showing the overlap of fluorescence from the retrograde tracer depicting BLA projectors and the fluorophore tdTomato depicting NT populations. (c) Quantification of overlap between NT expressing and BLA projecting neurons as a fraction of the NT population. (d) Schematic of viral-mediated CRISPR-Cas9 Nt gene knockout in each NT source. (e) Confocal images from a representative brain slice showing fluorescence from in situ hybridization of Nt mRNA and Vglut2 mRNA from the control and experimental groups. (f) Experimental design to test the acquisition of reward and punishment learning upon depletion of Nt gene. (g) Mice with PVT:Nt CRISPR-cKO showed a significantly higher port entry probability in last session of tone-sucrose conditioning, while MGN:Nt and vHPC:Nt CRISPR-cKO did not show significant changes compared to their cage-mate controls (P=0.9195, P=0.3976, and *P=0.0148, respectively for MGN, vHPC, and PVT. For CRISPR and control, N=18 and 17, N=11 and 9, and N=15 and 13, respectively for MGN, vHPC, and PVT). (h) CRISPR-cKO in none of the regions affected tone-shock acquisition in second session of tone-shock conditioning (P=0.4122, P=0.7818, and P=0.5751, respectively for MGN, vHPC, and PVT. For CRISPR and control, N=14 and 12, N=8 and 9, and N=8 and 9, respectively for MGN, vHPC, and PVT). Two-tailed unpaired t-test was used for all statistical tests. N denote number of mice in each group. Error bars and solid shaded regions around the mean indicate s.e.m. Shaded regions in panel g indicate the analysis windows used for statistical tests.
