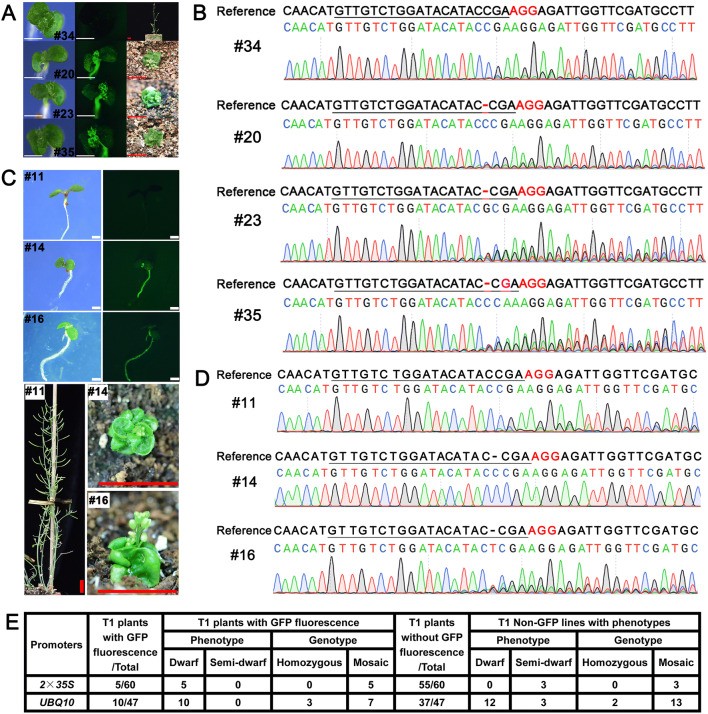
Fig. 3.
Cas9-P2A-GFP edited the genome efficiently. A, C The fluorescence intensity of transgenic plants correlated with the phenotype resulting from BRI1 editing. 4-day-old transgenic T1 plants were photographed using a Leica fluorescence stereoscope (A, left and middle; C, upper). The seedlings were then grown in the soil, and their phenotypes were recorded after several weeks. White bar = 1 mm. Red bar = 1 cm. Cas9-P2A-GFP was driven by the double CaMV 35S promoter (A) or the UBQ10 promoter (C). B, D Sanger sequencing results of representative T1 transgenic lines in panels A and C. BRI1 sgRNA sequences are underlined, and protospacer-adjacent motif (PAM) sequences are labeled in red. E Comparison of the editing efficiency of the 35S and UBQ10 promoters driving Cas9-P2A-GFP. The homozygous genotypes were based on Sanger sequencing in the T1 generation and were not confirmed in the T2 generation, since the T1 dwarf plants were sterile