T-cell acute lymphoblastic leukemia (T-ALL) is a rare subtype of ALL generally considered to behave less favorable than B-cell ALL, in particular when it displays an immature phenotype (early T-cell precursor [ETP]-ALL). It has a heterogeneous genetic background, characterized by the nonrandom involvement of multiple genes, which concur to delineate specific leukemogenic pathways.1 However, although recurrent alterations of transcription factors are useful to assign the leukemia to distinct genetic subgroups, and a number of oncogenes/suppressors have been extensively investigated, the leukemogenic activity of uncommon events remains to be elucidated. This is especially true for ETP-ALL cases.1
Eosinophilia is a typical presenting feature of a variety of hematological myeloid and lymphoid diseases. Eosinophils may be part of the neoplastic clone or represent a reactive cell population. Clonal eosinophils are found in acute myeloid leukemias (AMLs), characterized by RUNX1::RUNX1T1 or CBFB::MYH11, in chronic myeloid leukemia, systemic mastocytosis, and myeloid/lymphoid neoplasms with eosinophilia and PDGFRA, PDFGRB, FGFR1, JAK2, FLT3, or ETV6::ABL1 fusions.2 On the other hand, eosinophilia may be a reactive event due to increased production of growth factors, such as IL5 and IL3, in the classical form of Hodgkin disease (HD), T- and B-cell lymphomas, and histiocytic neoplasms. Interestingly, WHO recognizes a very rare subtype of B-cell acute lymphoblastic leukemia (B-ALL) with reactive eosinophilia, marked by the t(5;14)(q31;q32) translocation, in which constitutive IL3 activation occurs because of its juxtaposition nearby IGH regulatory sequences.2
Integration of our CI-FISH molecular cytogenetics assay3 with gene expressions studies, on 159 prospectively recruited T-ALLs (Suppl. Table S1), identified a unique case of immature T-ALLs with bone marrow (BM) and peripheral blood (PB) eosinophilia, in which PIM1 activation was caused by a previously undescribed t(5;7)(q31;q21)/CDK6::IL3.
The patient (index case), a 36-year-old man, was referred to the Hematology Department for generalized lymphadenopathy (Suppl. Material). Lymph node (LN) structure was subverted by an ancillary cell population with Langerhans morphology and phenotype, and low proliferative index. In a few areas, CD34+, TDT+, CD43+, MPO+/−, and CD7+ medium size cells, with scarce cytoplasm, cohesive growth, fine chromatin and inconspicuous nucleoli, were scattered. There were a few eosinophils. The PB cell count showed moderate leukocytosis (14.5 × 109) with eosinophilia (3.5 × 109), the presence of immature cells (0.8 × 109), and thrombocytopenia (119 × 109). BM was diffusely infiltrated by medium sized agranular blasts, with high nucleus/cytoplasmic ratio, basophilic cytoplasm, irregular nuclear membrane, disperse chromatin, and nucleoli. Immunophenotype was consistent with a diagnosis of near-ETP-ALL. The patient was treated according to the GIMEMA LAL1913 (NCT02067143) and, after achieving hematologic and cytogenetic remission (time point TP2), showed an increase of leukemic blasts (pre-hematopoietic stem cell transplantation [HSCT]). Hence, he received hematopoietic stem cells from an HLA-identical brother and achieved the molecular remission (Suppl. Material; Suppl. Table S2).
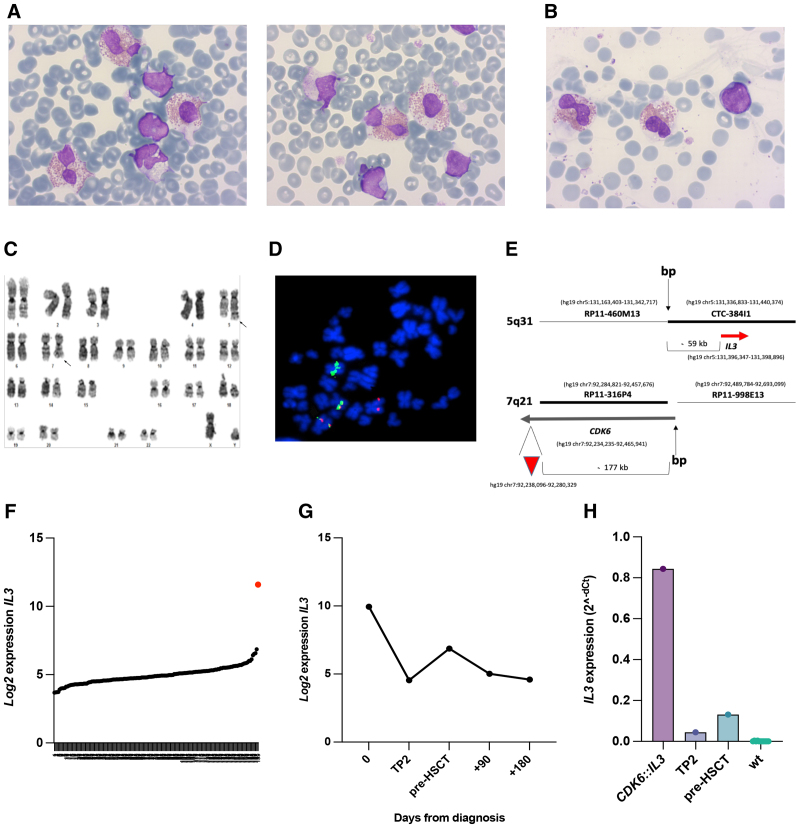
Interphase and metaphase fluorescence in situ hybridization, conducted on diagnostic PB samples, detected a previously unknown CDK6::IL3 rearrangement which was shown to cause the displacement of the IL3 gene from chromosome band 5q31 to der(7), where it located ~240 Kb apart from the CDK6 enhancer (position: hg19 position 92,238,096–92,280,329) (Figure 1C–E; Suppl. Results). The rearrangement was also demonstrated in 5%–15% of LN cells (Suppl. Results).
Figure 1.
Morphological features, conventional and molecular cytogenetics, and expression of IL3 in the index case with t(5;7)(q31;q21)/CDK6::IL3. (A and B) Leukemic blasts and eosinophils in the peripheral blood (A) and bone marrow (B) (May-Grunwald Giemsa stain) (Microscope Zeiis, magnification 100×; Camera AXIOCAM 105 color; Software ZEN Imaging Software 2.3); (C) Representative G-banded karyotype of the index case. Arrows indicate chromosomes 5 and 7 involved in reciprocal t(5;7)(q31;q21) (Microscope Olympus BX61, magnification 100×; Camera GENASIS Scanning System, ASI; Software GENASIS Band View, ASI); (D) Double color double fusion FISH experiment on an abnormal metaphase shows two fusion signals on der(5) and der(7), 1 orange signal, on normal chromosome 5, and 1 green signal on normal chromosome 7 (Microscope Olympus BX61, magnification 100×; Camera JAI progressive scan; Software CytoVision [Leica Microsystem]); (E) Schematic representation of DNA clones used for the CDK6::IL3 dual-color fusion assay, and their position relative to CDK6 and IL3. The red arrowhead indicates the position of the CDK6 enhancer; (F) A graph showing the expression levels of IL3 in our cohort of 159 T-ALL cases (RNA microarray data). The red dot corresponds to the index case with the CDK6::IL3 rearrangement; (G) Index case: longitudinal study of IL3 expression levels on samples taken at diagnosis and at different time points during therapy and after HSCT (see also Suppl. Table S1); (H) Validation of RNA microarray data by quantitative RT-PCR: IL3 expression in the index case, at diagnosis (CDK6::IL3) and during treatment (time points TP2 and pre-HSCT), and in 11 ETP-ALL cases without CDK6::IL3 (wt). ASI = applied spectral imaging; ETP-ALL = early T-cell precursor acute lymphoblastic leukemia; HSCT = hematopoietic stem cell transplantation; T-ALL = T-cell acute lymphoblastic leukemia.
As suggested by CDK6::IL3, RNA microarray showed that IL3 was overexpressed in the index case compared with the 158 cases without rearrangement (fold change +101.6, FDR P value 1.25 × 10−18) (Figure 1F). Indeed, the case belonged to the fourth quartile (Suppl. Table S3) and showed the highest level of IL3 (Suppl. Figure S1). Interestingly, an intermediate level of IL3 expression was detected in LN sections, with ~5%–15% of leukemic cell infiltration, while IL3 expression in the BM decreased during treatment, confirming the close correlation with the leukemic clone (Figure 1G). RNA microarray data were validated by qRT-PCR at 3 time points, that is, diagnosis, TP2, and pre-HSCT (Figure 1H).
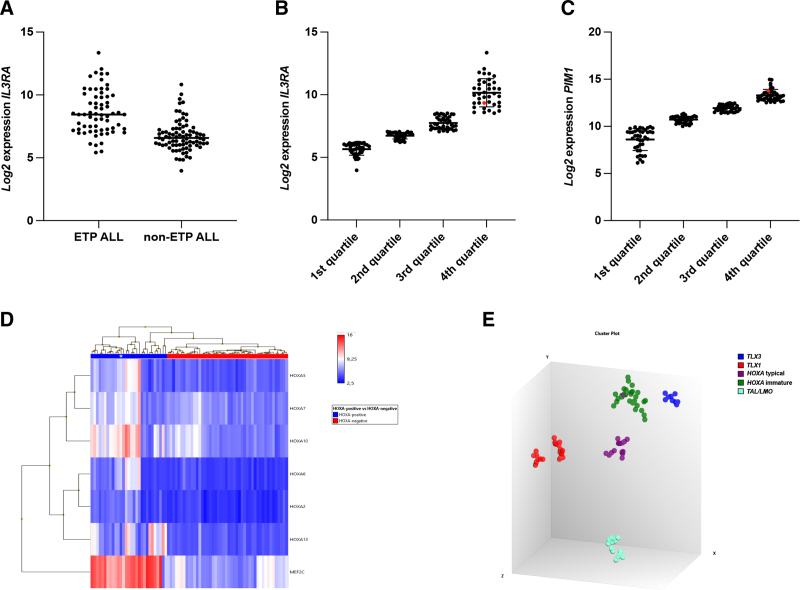
In hematopoietic cells, IL3 stimulates cell cycle progression and differentiation while inhibiting apoptosis.4 The IL3 regulatory signal is achieved through its binding with the IL3RA, encoding for the Interleukin 3 receptor subunit alpha, also known as CD123 antigen. High levels of IL3RA are typically found in hairy cell leukemia, HD, blastic plasmacytoid dendritic neoplasms, AML, and B-cell ALL.5 In T-ALL, IL3RA expression appears to be restricted to pediatric and adult pro-/ETP-ALL cases.6 Consistent with these data, we found that immature T-ALL in our cohort had the highest levels of IL3RA (Figure 2A) with 83% of cases (54/65) in the fourth (=29 case) and third (=25 case) quartiles (Suppl. Table S4). Notably, the CDK6::IL3 positive case also belonged to the fourth quartile (range: 8.5–13.35; median sample signals 9.95) (Figure 2B; Suppl. Figure S2).
Figure 2.
Gene expression studies. (A) IL3RA expression in ETP-ALL and non-ETP-ALL cases; (B) Distribution of 159 T-ALL cases into quartiles according to IL3RA expression. The index case belongs to the fourth quartile (red dot); (C) Distribution of 159 T-ALL cases into quartiles according to PIM1 expression. The index case belongs to the 4th quartile (red dot); (D) The index case (white asterisk) shows high expression levels of MEF2C and HOXA, and (E) clusters with HOXA-positive T-ALL with an immature phenotype (HOXA immature) (green dot marked with purple strokes). ASI = applied spectral imaging; ETP-ALL = early T-cell precursor acute lymphoblastic leukemia; T-ALL = T-cell acute lymphoblastic leukemia.
Altogether our findings suggest that CDK6::IL3 might have exerted the same functional consequences of the IGH::IL3 rearrangement in B-ALL,2 mirroring the transcriptional activation of IL3 by chromosomal translocation, with an increase of circulating reactive eosinophils.2 In both subtypes of leukemia, IL3 could play a dual action on cell proliferation, that is, a paracrine effect on eosinophils and an autocrine effect on leukemic blasts, as already described in other solid and hematologic cancers.7,8 Interestingly, a possible paracrine effect was also observed in our case at the LN level, where there is an overwhelming population of Langerhans cell. Findings of partial IL3RA antigen expression (Suppl. Results and Suppl. Figure S3) and an undetectable serum level of IL3 (Suppl. Results and Suppl. Tables S5 and S6), support the paracrine and autocrine activity of IL3, which may have caused IL3RA downregulation,9 and failure to release IL3 into PB.10 It is worth mentioning that an autocrine production pattern has been reported in T-ALL for IL7, which, as a consequence of promoter hypomethylation, is secreted by leukemic cells contributing to the development of leukemia.,11
It is worth noting that the IL3-IL3RA signaling is transduced through the JAK/STAT pathway, which controls the transcription of a number of target genes, among which PIM1 is a known oncogene in T-ALL, and more widely in human cancers.12 PIM1 is typically upregulated in T-ALL with gain-of-function mutations of IL7R or JAK/STAT members (20%–30% of T-ALL), haploinsufficiency/inactivation of the negative regulators PTPN2 and PTPRC (6%), and in rare cases harboring the t(6;7)(p21;q34) translocation.1 Our case report adds the CDK6::IL3 rearrangement to the mechanisms underlying PIM1 overexpression (Figure 2C; Suppl. Figure 4, and) suggest that a number of yet unknown alterations may cause PIM1 cis- or trans- activation.
Regardless of the mechanism, high levels of PIM1 are mainly associated with genomic abnormalities of homeobox deregulated subgroups (HOXA, TLX1, and TLX3; Suppl. Table S7), and with an immature phenotype.13 Although no genomic abnormalities were found to classify the index case into one of the major genetic subgroups, RNA microarray showed high levels of MEF2C, as expected in immature T-ALL, and of a number of HOXA genes (Figure 2D). Specifically, our probe set sub-grouped T-ALLs with rearrangements of the HOXA genes, or of genes trans-activating HOXA, according to the immunophenotype, that is, HOXA-positive typical (early, cortical, and mature) and HOXA-positive immature (ETP and near-ETP-ALL) (Figure 2E and Suppl. Results). In agreement with the near-ETP phenotype, our index case belongs to the latter subtype (Figure 2E).
Seeking for concurrent abnormalities that co-operate with the JAK/STAT signaling, we found mutations that are typical of immature T-ALL,14 that is, DNMT3A c.2644C>T p.Arg882Cys (VAF 25.9%), which is recurrent in AML, and two frameshifts at ETV6 gene, the c.433_434insTCTTTTG p.Glu145Valfs*3 (VAF 24.9%) and the c.526dupA p.Ile176Asnfs*2 (VAF 24.8%). SNPa revealed a unique event, consistent of a ~8Mb region of loss at 8q23q24, previously described in a case of AML (Suppl. Results).15
Our comprehensive study led to the identification of a unique case of T-ALL in which the t(5;7)(q31;q21)/CDK6::IL3 rearrangement represents a novel mechanism of IL3 transcriptional activation and, consequently, eosinophilia. Additionally, CDK6::IL3 is a new leukemogenic event to unlock the JAK/STAT signaling and upregulating downstream targets, including PIM1. Therefore, it might serve as a predictive marker of sensitivity to IL3RA and/or of PIM1 inhibitors. The IL3 gene study in the diagnostic work-up of hematologic malignancies with eosinophilia is strongly recommended.
AUTHOR CONTRIBUTIONS
VP, SA, VB, MQ performed and analyzed FISH, SNPa, RNA microarray, and gene expression studies (RTq-PCR); CM, VB, and FP performed and analyzed targeted next generation sequencing; FG, AB, and FC provided all clinical and hematological data; PR, MP, ES, EF, and LR performed and analyzed morphology, flow cytometry, histology, and/or immunohistochemistry; RLS conceived the study and wrote the paper; CM supervised and wrote the paper. All authors approved the paper.
DISCLOSURES
The authors have no conflicts of interest to disclose.
Supplementary Material
Footnotes
VP, VB, FG, CM, and RLS share co-authorship.
Supplemental digital content is available for this article.
REFERENCES
- 1.Bardelli V, Arniani S, Pierini V, et al. T-cell acute lymphoblastic leukemia: biomarkers and their clinical usefulness. Genes (Basel). 2021;12:1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian RP, et al. International consensus classification of myeloid neoplasms and acute leukemia: integrating morphological, clinical, and genomic data. Blood. 2022;140:1200–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Starza R, Pierini V, Pierini T, et al. Design of a comprehensive fluorescence in situ hybridization assay for genetic classification of T-cell acute lymphoblastic leukemia. J Mol Diagnostics. 2020;22:629–639. [DOI] [PubMed] [Google Scholar]
- 4.Blalock WL, Weinstein-Oppenheimer C, Chang F, et al. Signal transduction, cell cycle regulatory, and anti-apoptotic pathways regulated by IL-3 in hematopoietic cells: possible sites for intervention with anti-neoplastic drugs. Leukemia. 1999;13:1109–1166. [DOI] [PubMed] [Google Scholar]
- 5.Munoz L, Nomodedéu JF, López O, et al. Interleukin-3 receptor alpha chain is widely expressed in hematologic malignancies. Haematologica. 2001;86:1261–1269. [PubMed] [Google Scholar]
- 6.Testa U, Riccioni R, Diverio D, et al. Interleukin-3 receptor in acute leukemia. Leukemia. 2004;18:219–226. [DOI] [PubMed] [Google Scholar]
- 7.Aldinucci D, Poletto D, Gloghini A, et al. Expression of functional interleukin-3 receptors on Hodgkin and Reed-Sternberg cells. Am J Pathol. 2002;160:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang X, Lopez A, Holyoake T, et al. Autocrine production and action of IL-3 and granulocyte colony-stimulating factor in chronic myeloid leukemia. Proc Natl Acad Sci USA. 1999;96:12804–12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steelman LS, Algate PA, Blalock WL, et al. Oncogenic effects of overexpression of the interleukin-3 receptor on hematopoietic cells. Leukemia. 1996;10:528–542. [PubMed] [Google Scholar]
- 10.Roodman GD, Kurihara N, Ohsaki Y, et al. Interleukin 6: a potential autocrine/paracrine factor in Paget’s disease of bone. J Clin Invest. 1992;89:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buffière A, Uzan B, Aucagne R, et al. T-cell acute lymphoblastic leukemia displays autocrine production of Interleukin-7. Oncogene. 2019;38:7357–7365. [DOI] [PubMed] [Google Scholar]
- 12.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11:23–34. [DOI] [PubMed] [Google Scholar]
- 13.La Starza R, Messina M, Gianfelici V, et al. High PIM1 expression is a biomarker of T-cell acute lymphoblastic leukemia with JAK/STAT activation or t(6;7)(p21;q34)/TRB@-PIM1 rearrangement. Leukemia. 2018;32:1807–1810. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toft-Petersen M, Kjeldsen E, Nederby L, et al. A novel del(8)(q23.2q24.11) contributing to disease progression in a case of JAK2/TET2 double mutated chronic myelomonocytic leukemia. Leuk Res Reports. 2014;3:94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.