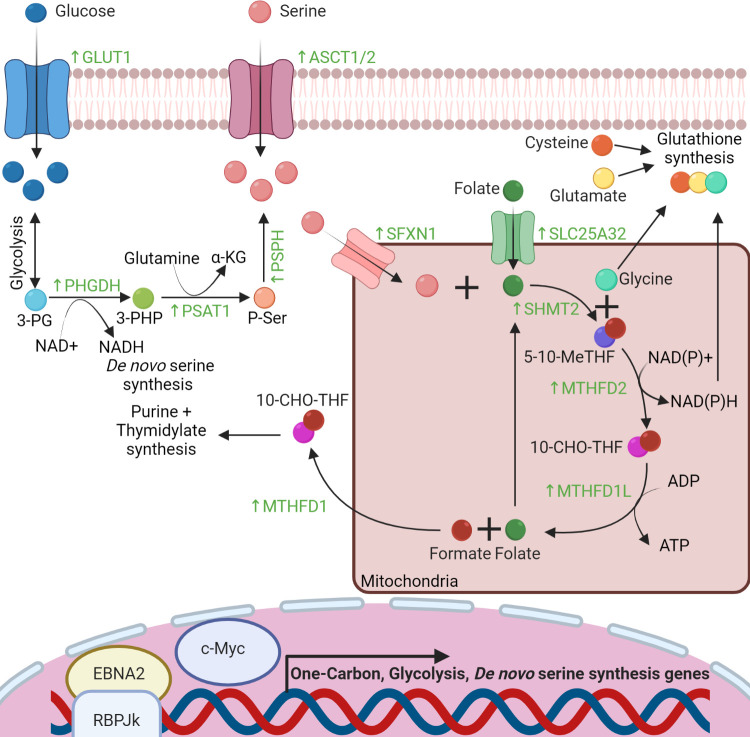
Fig 2 is incorrect. The labels for SLC25A32 and SFXN1 are swapped. The authors have provided a corrected version of Fig 2 here.
Fig 2. EBV-driven 1C pathway drives nucleotide and glutathione synthesis.
EBNA2 and MYC induce the 1C, glycolysis, and de novo serine synthesis pathways in B cells. They also up-regulate plasma membrane abundance of the GLUT1 and ASCT1/2 transporters to increase glucose and serine import, respectively. Serine pools are further expanded by the de novo serine synthesis pathway, which metabolizes the glycolysis product 3-PG into serine and generate NADH and αKG byproducts. EBV also supports mitochondrial 1C metabolism via up-regulation of the SLC25A32 serine and SFXN1 folate transporters. Mitochondrial 1C metabolism converts serine into glycine, NADPH, and a serine-derived carbon unit (red ball), which is shuttled into the cytosol for use in purine and thymidylate biosynthesis. The carbon-loaded 1C folate carriers 5-10-meTHF and 10-CHO-THF are shown. 1C, one-carbon; 3-PG, 3-phosphoglycerate; 3-PHP, 3-phosphohydroxypyruvate; P-ser, 3-phosphoserine; 5-10-meTHF, 5-10-methylenetetrahydrofolate; 10-CHO-THF, 10-formyl THF; EBNA2, Epstein–Barr nuclear antigen 2; EBV, Epstein–Barr virus.
Reference
- 1.Burton EM, Gewurz BE (2022) Epstein–Barr virus oncoprotein–driven B cell metabolism remodeling. PLoS Pathog 18(2): e1010254. 10.1371/journal.ppat.1010254 [DOI] [PMC free article] [PubMed] [Google Scholar]