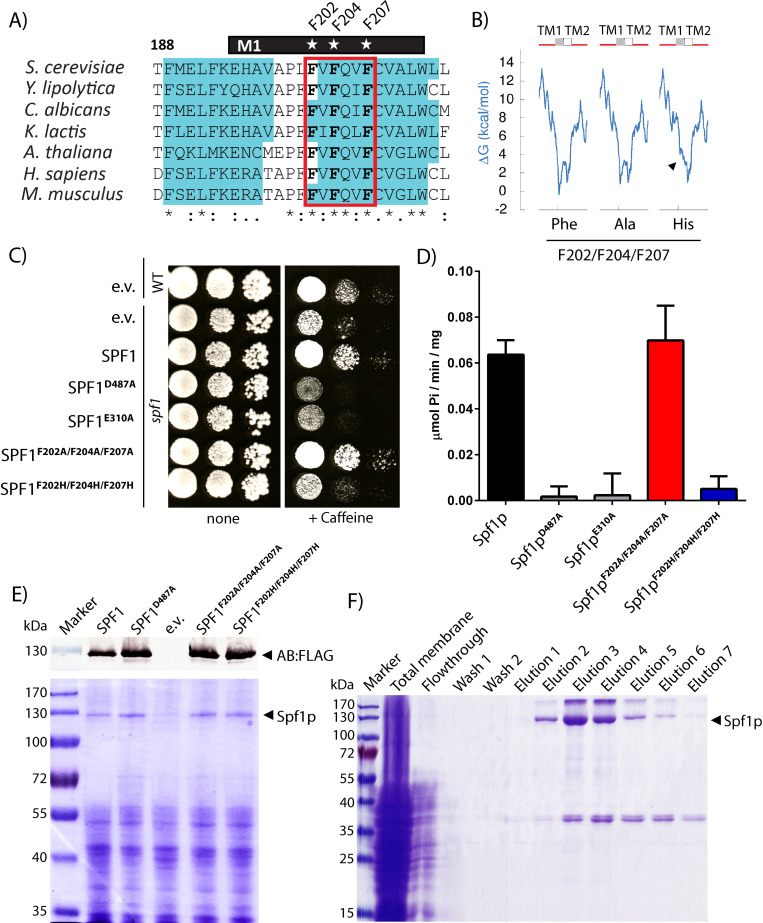
Fig 5. Viability of Spf1p and M1 mutated Spf1p.
A) Alignment of M1 region of Spf1p and similar P5A ATPase sequences. Conserved residues F202, F204 and F207 indicated by star and predicted α-helical structure shown in blue. B) Calculated ΔG (kcal/mol) for folding of M1 and M2 in Spf1p and M1 mutated spf1p. Introduction of histidines, but not alanines, increase the calculated energetic requirement for proper folding into the α-helical structure. C) Complementation in spf1 deletant cells using the expression constructs used in this study. The catalytic dead mutations D487A and E310A and the empty vector was used as a control. Complementation was tested on 10 mM caffeine plates as described in [12]. D) Specific activity of purified Spf1p mutants in mixed YPL/OG micelles. All experiments are performed as biological replicates and technical triplicates (n = 6) and is reported as the average value of these with error bars indicating the standard error. E) Expression analysis of the constructs used in this study, SDS-PAGE show expression of a ~130 kDa band corresponding to Spf1 and M1 mutated Spf1p. A western blot against the FLAG-tag using Anti-FLAG AB is shown for reference. F) SDS-PAGE gels of a typical purification showing relatively pure and homogenous preparations comparable to those presented in [12]. Fractions containing Spf1p were pooled for recovery of purified Spf1p.