Figure 5. PARLSkd3 is functional at low ATP concentrations.
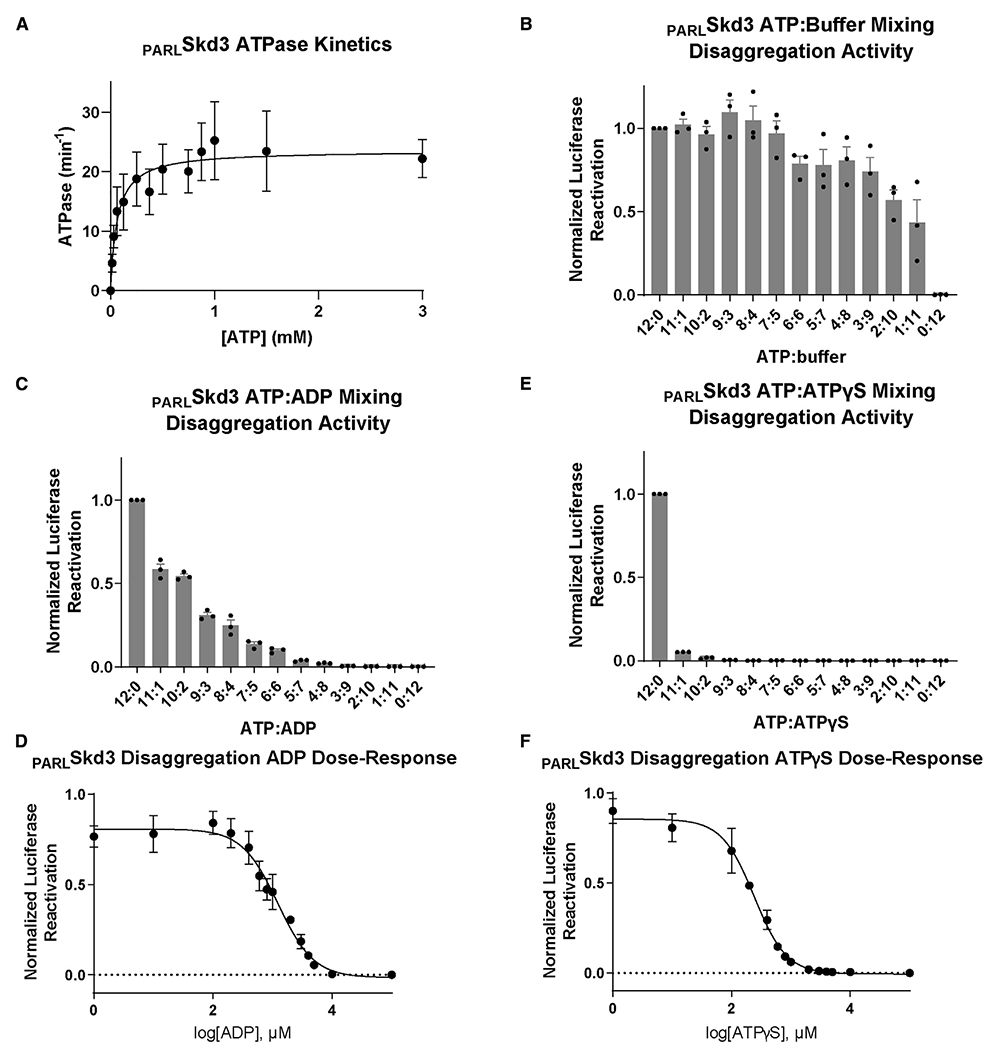
(A) Michaelis-Menten plot of PARLSkd3 ATPase activity. Vmax ~ 23.6 min−1 and KM ~ 64.6 μM (n = 3, values are means ± SEM).
(B) Luciferase disaggregase activity of PARLSkd3 at various ATP:buffer ratios. ATP concentrations used were 12:0 (5 mM), 11:1 (4.58 mM), 10:2 (4.17 mM), 9:3 (3.75 mM), 8:4 (3.33 mM), 7:5 (2.92 mM), 6:6 (2.5 mM), 5:7 (2.08 mM), 4:8 (1.67 mM), 3:9 (1.25 mM), 2:10 (0.83 mM), 1:11 (0.42 mM), and 0:12 (0 mM). Disaggregase activity was normalized to PARLSkd3 plus ATP (n = 3, individual data points shown as dots, bars show mean ± SEM).
(C) Luciferase disaggregase activity of PARLSkd3 at various ATP:ADP ratios where the total nucleotide concentration was 5 mM. Disaggregase activity was normalized to PARLSkd3 plus ATP (n= 3, individual data points shown as dots, bars show mean ± SEM).
(D) Luciferase disaggregase activity of PARLSkd3 at a constant concentration of ATP (5 mM) and increasing ADP concentrations. Disaggregase activity was normalized to PARLSkd3 plus ATP (n = 3, values are means ± SEM).
(E) Luciferase disaggregase activity of PARLSkd3 at various ATP:ATPγS ratios where the total nucleotide concentration was 5 mM. Disaggregase activity was normalized to PARLSkd3 plus ATP (n = 3, individual data points shown as dots, bars show mean ± SEM).
(F) Luciferase disaggregase activity of PARLSkd3 at a constant concentration of ATP (5 mM) and increasing ATPγS concentrations. Disaggregase activity was normalized to PARLSkd3 plus ATP (n = 3, data shown as mean ± SEM).
