Abstract
A study was conducted to determine the prevalence and drug resistance of Escherichia coli present in urinary tract infected patients and hospital drinking water. A total of eighty urine samples from clinically suspected patients and thirty tap water samples from hospital vicinity were collected and analyzed for the presence of E. coli. The isolates were preliminary identified based on morphological characteristics, biochemical test and further confirmed by polymerase chain reaction (PCR) using uidA primer. Isolates were subjected to antibiogram studies and analyzed for the presence of drug resistance (ESBL blaCTX-M-15, tetA, and TMP-SMX dfrA1) and pathogenicity associated pyelonephritis-associated pili (PAP) and Heat-labile (LT) toxin genes. Urine samples 19/80 (23.75%) and water samples 8/30 (26.7%) were found contaminated with E. coli. It was found that 12/19 (63%) bacterial isolates were extended spectrum beta-lactamase (ESBL) producers in clinical and 6/8 (75%) in water isolates whereas tetracycline resistance in clinical and water isolates was 11/19 (58%) and 6/8 (75%), respectively. The trimethoprim resistance gene was confirmed in 12/19 (63%) in clinical and 2/8 (25%) in water isolates. All the clinical and water isolates were found carrying pili PAP gene. It was concluded that the presence of drug resistant and pathogenic E. coli in clinical and water samples is extremely alarming for public health due to cross contamination and bacterial transfer from clinical samples to water and vice versa.
1. Introduction
Antibiotics are essential tool against pathogenic bacterial infections but the rapid emergence of antibiotic resistant pathogens is currently creating an alarming situation [1]. Drug resistance is a global public health concern, associated with more than 700,000 deaths annually [2]. This growing threat necessitates the action to prevent the emergence of new resistant bacterial strains and the spread of existing ones in the environment from humans and animals [3]. Antibiotic resistant bacteria have been found in wastewater, surface water, and drinking water. Water is regarded to be the most important medium for the transmission of antibiotic resistance. This is especially true in low-income nations, where lack of sanitation and hygiene practices aggravates the problem [4]. Hospital sanitation water is associated with the presence of antibiotic residues and resistant microorganisms which are delivered into the aquatic ecosystem [5]. Each year, hospitals in Pakistan are roughly generating 25,000 tons of wastes with antibiotic residues ranging from 1.4 g/L to 236.6 g/L, which is thrown into surface water untreated [6]. Due to lack of appropriate management, drinking water is regularly mixed with sewage water and contaminated hospital effluent that poses a serious threat to the human population. In such conditions, the presence of antibiotic resistant bacteria increases the possibility of horizontal genetic transmission between microbes, leading to the emergence of multidrug resistant bacteria [7, 8]. E. coli can easily acquire drug resistance genes and is responsible for a variety of human diseases, including traveler's and infantile diarrhea, urinary tract infections, septicemia, hemolytic uremic syndrome, newborn meningitis, and pneumonia [9, 10]. E. coli is a commensal, opportunistic pathogen that is found in animals' gastrointestinal tracts as well as in the environment, such as in water and soil [11]. In this work, E. coli bacteria isolated from clinical and water sources were evaluated for drug resistance pattern and virulence potential. The aim of this study was to determine the prevalence of pathogenic and drug resistant hospital acquired E. coli from clinical and hospital water samples. The findings of this study will be useful globally in antibiotic treatment and monitoring the resistance patterns over time in developing and underdeveloped regions.
2. Methodology
2.1. Sampling
Urine samples (n = 80) were collected aseptically in sterile sampling containers from the patients having clinical signs such as pelvic pain, burning sensation during urination, high fever, and blood in urine. All the samples were collected using convenience random sampling technique from Bolan Medical Complex Hospital and Provincial Sandeman Hospital Quetta, city of Pakistan from April to December 2018, following all the safety procedures and protocols. Informed consent was obtained from all the participants.
Drinking tap water samples (n = 30) were collected from hospital vicinity aseptically using sterilized glass bottles. All the samples (clinical and water) were transported to Center for Advanced Studies in Vaccinology and Bacteriology, University of Balochistan, and during transportation, temperature (0–4°C) was maintained. The samples were immediately (within 6 h of collection) processed for the isolation of target pathogen E. coli.
2.2. Isolation and Identification
The clinical samples were streak directly over the surface of sterile MacConkey agar (MCA) (Oxoid, UK) plates, while the water samples were first enriched in lactose broth for 6–16 h at 37°C, then transferred to MCA, and incubated at 37°C for 24 h [12].
Suspected lactose fermenting pink colonies from MCA were transferred to Eosin Methylene Blue (EMB) agar (Oxoid, UK) and incubated at 37°C for 16–24 h [13]. Colonies with green metallic sheen and typically resembling E. coli were further characterized. Gram's staining, Catalase, Oxidase, Sulfide, Indole Motility, Methyl Red, Voges Proskauer, Simmon citrate utilization, and urease tests were performed for the biochemical conformation of the E. coli isolates [12].
2.3. Molecular Identification
The preliminary identified E. coli isolates were subjected to molecular identification using E. coli UidA primer that amplifies 147 bp gene size product [14]. Bacterial DNA was extracted by using the DNA Mini Kit (Qiagen), and polymerase chain reaction (PCR) was carried out by the use of specific oligonucleotide primers. PCR mixture (25 μl) was comprised of master mix (12.5 μl), molecular grade water (7.5 μl), 1 μl of primer (forward and reverse), and 3 μl of DNA template. Using a thermocycler (Bio-Rad, USA), PCR was carried out (Table 1). PCR products were analyzed by electrophoresis in 1.2% agarose gel, and visualization of bands was done in transilluminator (Dolphin View-Wealtec, USA) under UV light.
Table 1.
Details of primers and their respective polymerase chain reaction conditions.
| Gene | Size | Polymerase chain reaction protocol | Sequence of gene |
|---|---|---|---|
| UidA | 147 bp | Initial denaturation was carried out for 7 min at 95°C followed by total 35 cycles of 30 sec each at 95°C; annealing for 30 sec at 55°C; extension for 30 sec at 72°C, followed by final extension at 72°C, for 7 min | F-TGGTAATTACCGAC GAAAACGGC R-ACGCGTG GTTACAGTCTTGCG |
|
| |||
| blaCTX-M-15 | 996 bp | Initial denaturation for 4 min at 96°C followed by total 34 cycles of 20 sec each at 95°C; annealing for 20 sec at 56°C; 60 sec at 72°C, followed by final extension at 72°C, for 5 min | F-CACACGTGGAATTTAGGGACT R-GCCGTCTAAGGCGATAAACA |
|
| |||
| tetB | 634 bp | Initial denaturation for 3 min at 95°C followed by total 35 cycles of 1 min each at 94°C; annealing for 90 sec at 56°C; 60 sec at 72°C, followed by final extension at 72°C, for 8 min | F-CCTCAGCTTCTCAACGCGTG R-GCACCTTGCTGATGACTCTT |
|
| |||
| TMP-SMX dfrA1 | 367 bp | Initial denaturation for 3 min at 95°C followed by total 35 cycles of 1 min each at 94°C; annealing for 90 sec at 45°C; 60 sec at 72°C, followed by final extension at 72°C, for 8 min | F-GGAGTGCCAAAGGTGAACAGC R-GAGGCGAAGTCTTGGGTAAAAAC |
|
| |||
| PAP | Initial denaturing 3 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 58°C, 1 min at 72°C followed by final extension of 5 min at 72°C | F-AACCTGGCTTACGCAACTGTACCC GT R-CTG CAA AAT CAT GGA T |
|
|
| |||
| Heat-labile toxin | Initial denaturing 3 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C and final extension for 5 min at 72°C | F-GCACACGGAGCTCCTCAGTC R-TCCTTCATCCTT TCA ATG GCT TT |
|
2.4. Antibiogram of Isolates
Antibiogram of E. coli isolated was determined through Kirby-Bauer disk diffusion procedure using Mueller Hinton agar (MHA) (Oxoid, UK), following Clinical Laboratory and Standard Institute (CLSI) guidelines [15]. Overnight grown culture was assorted in 1 mL of sterile saline solution equating the turbidity to 0.5 McFarland standard followed by swabbing on MHA [16]. Twelve frequently prescribed antibiotics were impregnated on the surface of MHA comprising of tetracycline (30 μg), gentamycin (10 μg), chloramphenicol (30 μg), trimethoprim-sulphamethoxazole (25 μg), ciprofloxacin (5 μg), ceftazidime (30 μg), amikacin (30 μg), cefixime (5 μg), Imipenem (10 μg), amoxicillin (30 μg), amoxicillin + Clavulanic Acid (30 μg), and cefotaxime (30 μg). After incubation of 24 h at 37°C, diameter of inhibition zone was measured.
2.5. ESBL Conformation Test
The double disc synergy test was done on MHA to determine extended spectrum beta-lactamase (ESBL) producing isolates. Ceftazidime and Cefotaxime discs were spaced 15 mm away from an amoxicillin-clavulanic acid (20/10 μg) disc in the plate's middle [17].
2.6. Molecular Detection of Drug Resistance Genes
Genes responsible for drug resistances against selected antibiotics were determined using gene specific primers for the targeted genes including ESBL blaCTX-M-15 gene that amplifies 996 bp gene size product [13], tetracycline tetB gene that amplifies 634 bp gene size product [18], and TMP-SMX dfrA1 resistance gene that amplifies 367 bp gene size product [19].
By using thermocycler, PCR mixture (25 μl) was used to amplify ESBL blaCTX-M-15 gene, tetB gene, and TMP-SMX dfrA1 (Table 1).
2.7. Molecular Detection of Pathogenicity-Related Genes
Two virulence genes PAP [20] and Heat-labile toxin [21] were targeted to determine the pathogenicity potential of the isolates by PCR. By using thermocycler, PCR mixture (25 μl) was used to amplify PAP gene (initial denaturing 3 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 58°C, 1 min at 72°C followed by final extension of 5 min at 72°C) and heat-labile toxin gene (initial denaturing 3 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C and final extension for 5 min at 72°C).
2.8. Statistical Analysis
Statistical calculation of the data was done by using Microsoft Excel. Chi-square goodness-of-fit test was used for the determination of difference in resistance. p values < 0.05 were considered statistically significant.
3. Results and Discussion
A total 110 sample comprising 80 urine and 30 tap water samples were analyzed for the detection of E. coli. Out of total 110 samples, 25% (27/110) were found contaminated with E. coli, whereas 23.75% (19/80) urine sample was positive for the presence of the target pathogen, while 26.7% (8/30) water sample was confirmed with the presence of E. coli through conventional and molecular methods (PCR) targeting uidA gene (Figure 1).
Figure 1.
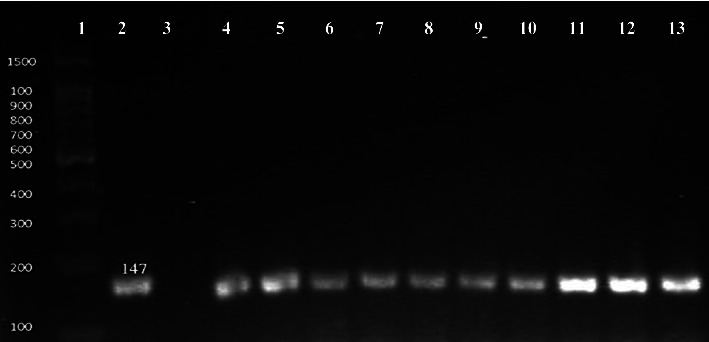
Molecular conformation of E. coli by uidA gene. Number 1: 100 bp DNA ladder; number 2: positive control; number 3: negative control; number 4 to 13: positive samples.
Due to frequent and unnecessary use of antibiotics, E. coli is becoming drug resistant, making it a superbug and a huge public health risk. Antibiotic susceptibility of E. coli isolates must be determined for the proper and precise prescriptions. If proper management and precautions have not been taken for the prevention of drug resistance, the current death toll 700,000 per year can be increased to ten million mortality rate every year throughout the globe by year 2050 [22]. Drug resistance is a barrier to universal health care and achieving sustainable development goals in the areas of food security, clean water, sanitation, and health [23].
Contrary to our finding, Talukdar et al. [13] reported 63% prevalence of E. coli, in household water, which was relatively high compared to our findings. Similar to our study, Shimpoh et al. [24] used uidA primer to identify E. coli strains from clinical samples collected from Japanese patients and reported a high detection rate of 51%.
3.1. Antibiogram
Twelve commercially available antibiotic discs were used to determine the antibiogram of the isolates. A majority of the isolates showed resistance to one or more antibiotics. Antimicrobial susceptibility test results revealed that few isolates showed susceptibility against chloramphenicol, amikacin, and imipenem while large number of E. coli isolates showed 100% (27/27) resistance to tetracycline, amoxicillin, amoxicillin-clavulanic acid, cefotaxime, and cefixime in clinical and water isolates (Figure 2).
Figure 2.
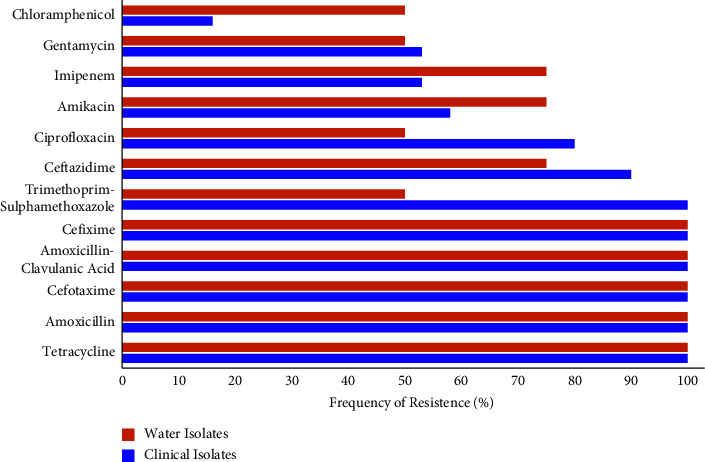
Drug resistance pattern in E. coli isolates from clinical and water samples.
In agreement with our results, previous reports (23, 24) documented that E. coli showed resistance against tetracycline and ceftazidime. Comparing with our work, research conducted in Bangladesh revealed that E. coli showed higher resistance against tetracycline (91.22%), trimethoprim (29.82%), and chloramphenicol 29% [25]. Another study demonstrated that E. coli was resistant to ampicillin (98%), tetracycline (23%), and trimethoprim-sulpha methoxazole (38%), respectively [26]. Contrary to our findings, previously a lower resistance in E. coli isolates, 45% tetracycline, 36% trimethoprim-sulfamthoxazole, 17% ciprofloxacin, 8% chloramphenicol, and 1% gentamycin was documented [13]. Scaria et al. [27] studied that all E. coli strains were found resistant to streptomycin (95%), tetracycline, ampicillin (47%), and kanamycin (91%).
Moges et al. [28] reported 100% ampicillin resistant and 38% tetracycline resistant E. coli isolated from hospital wastewater. They also reported high resistance of E. coli isolated from hospital wastewater to cefotaxime (38%), trimethoprim/sulfamethoxazole (38%), and cephalothin (23%). Tetracycline (21.45%), trimethoprim/sulfamethoxazole (18.56%), ampicillin (11.32%), ciprofloxacin (8.25%), amikacin (7.22%), gentamicin (5.15%), and cefotaxime (4.12%) resistance was found in E. coli isolates from contaminated water in Ghana [29]. These investigations have demonstrated that the habitat has a significant impact on E. coli antibiotic resistance due to exposure to various levels of drug residues in surrounding environment.
Out of 27 isolates, only 5 were susceptible to ceftazidime while other isolates showed resistance against various commercially available antibiotics. A research study conducted in Saudi Arabia reported that ESBL producing E. coli isolate demonstrated 100% resistance against amoxicillin/clavulanic acid and showed higher resistance rate against ceftazidime 100% and amikacin 54% comparing with present results 80% and 50%, respectively [30]. The current study suggested that the commercially available antibiotics such as amikacin, ceftazidime, and imipenem are potent against ESBL E. coli isolates.
3.2. Drug Resistance and Virulence Gene
The results for the identification of drug resistance genes revealed that ESBL blaCTX-M-15 gene was found in 63% (12/19) of clinical and 75% (6/8) of tap water isolates (Figure 3) followed by tetracycline resistance gene tetA 58% (11/19) in clinical and 75% (6/8) in tap water isolates (Figure 4). The TMP-SMX dfrA1 resistance gene was found in 63% (12/19) of clinical and 25% (2/8) of tap water isolates, respectively (Figure 5).
Figure 3.
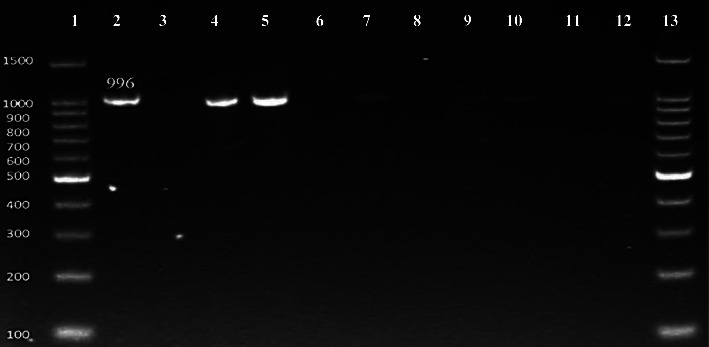
Molecular detection of ESBLs (blaCTX-M-15) gene (996 bp). Number 1 and 13: 100 bp DNA ladder; number 2: positive control; number 3, 6, 8, 11: negative samples; number 4, 5, 7, 9, 10, 12: positive samples.
Figure 4.
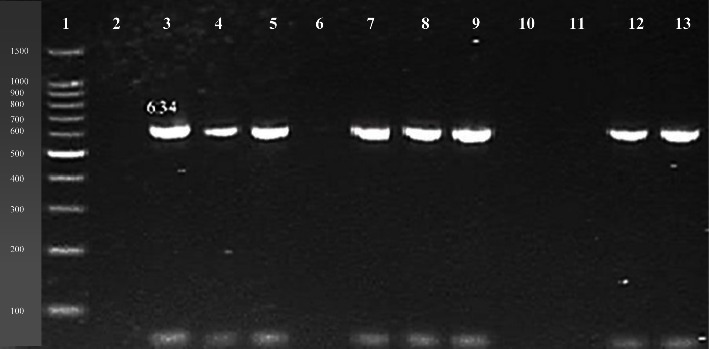
Molecular detection of tetB drug resistance gene (634 bp). Number 1: 100 bp DNA ladder; number 2, 6, 10, 11: negative samples; number 3: positive control; number 4, 5, 7, 8, 9, 12, 13: positive samples.
Figure 5.

Molecular detection of dfrA1 gene (367 bp). Number 1: 100 bp DNA ladder; number 2: positive control; number 3, 6, 8, 12: negative samples; number 4, 5, 7, 9, 10, 11, 13: positive samples.
In comparison to our findings, a high prevalence rate (66%) of bla CTX-M-15 ESBL gene was found in E. coli isolate from water samples [31]. Comparing with present work, a research work reported lower prevalence rate of CTX-M-15 ESBL gene (43%) in E. coli clinical isolates [13]. Lyimo et al. [32] studied the prevalence of blacCTX-M-19 and blaCTX-M-15 in E. coli from water samples and reported lower prevalence comparing with the current study. In a study conducted in Pakistan, blaCTX-M gene was found in 29% E. coli isolates from water sample. BlaSHV and blaTEM were shown to impart resistance to several antibiotics in their investigation, and they were the most widespread in E. coli, which contained the highest number of resistance genes, including blaCTXM, blaOXA, and blaNDM. [33].
Tetracycline (tetA, tetB, and tetC) and trimethoprim-sulfamethoxazole resistance genes were investigated by Ibekwe et al. [34] in E. coli. The results of their study revealed that 10 E. coli isolates were positive for tetB gene and 10 isolates were also positive for dfrA1 gene. Comparing with the present study, a research study conducted in Iran, reported that tetB (53.63%) and dfrA1 genes (36.84%) were present in PCR-confirmed E. coli isolates [25]. Another study documented that tet B and TMP-SMX dfrA1 gene were highly prevalent in E. coli from clinical samples [35].
The virulence gene PAP was found in all (100%) the confirmed E. coli isolates (Figure 6) while Heat-labile toxin gene was found absent in all isolates. In a similar study, it was reported that E. coli isolates tested for virulence gene PAP and Heat-labile toxin (LT) revealed 18% presence of PAP gene [36] and 12.7% LT gene detection in the isolates [37]. In another study conducted in South Africa by Nontongana et al. [26], 2% PAP gene and 47% LT gene presence were observed in the E. coli isolates. In comparison to the current study, literature variations in the prevalence of E. coli in clinical samples and antibacterial resistance might be due to different geographical regions, difference in the level of antibiotic exposure, and variation in E. coli strains. A detailed study by including larger sample size and collection of samples from different sampling sites around the region may provide more comprehensive findings.
Figure 6.
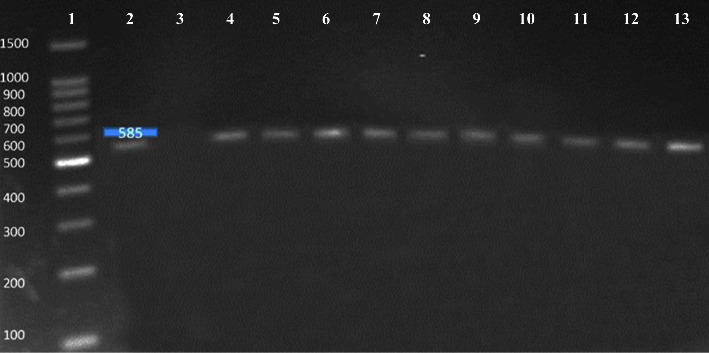
Molecular detection of PAP virulence gene (585 bp). Number 1: 100 bp DNA ladder; number 2: positive control; number 3: negative control; number 4 to 13: positive samples.
4. Conclusion
It was concluded in this study that E. coli bacteria isolated form clinical and hospital vicinity water were resistant (100%) to Tetracycline, trimethoprim-sulfamethoxazole, Cefizime, and amoxicillin. Furthermore, drug resistance (ESBL blaCTX-M-15, tetA, and TMP-SMX dfrA1) and pathogenicity associated PAP genes were also present in the bacterial isolates, which presented a serious health concern. Precautions are required to reduce the incidence of multidrug resistant E. coli strains among humans. In order to achieve this, strict quality control procedures should also be used to ensure that water and wastewater are properly treated.
Contributor Information
Ali Akbar, Email: aliakbar.uob@gmail.com.
Su Hlaing Chein, Email: suhlaing.shc08@gmail.com.
Data Availability
Major part of the data is presented in the manuscript, and the remaining data will be made available on reasonable request.
Ethical Approval
The study was conducted after the ethical approval from the Institutional Bioethics Committee, whereas the samples were collected after the patients' informed consent.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Gul Z., Akbar A., Leghari S. K. Elucidating therapeutic and biological potential of berberis baluchistanica ahrendt bark, leaf, and root extracts. Frontiers in Microbiology . 2022;13 doi: 10.3389/fmicb.2022.823673.823673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalungia A. C., Mwambula H., Munkombwe D., et al. Antimicrobial stewardship knowledge and perception among physicians and pharmacists at leading tertiary teaching hospitals in Zambia: implications for future policy and practice. Journal of Chemotherapy . 2019;31(7-8):378–387. doi: 10.1080/1120009x.2019.1622293. [DOI] [PubMed] [Google Scholar]
- 3.Rosa G., Huaylinos M. L., Gil A., Lanata C., Clasen T. Assessing the consistency and microbiological effectiveness of household water treatment practices by urban and rural populations claiming to treat their water at home: a case study in Peru. PLoS One . 2014;9(12) doi: 10.1371/journal.pone.0114997.e114997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker G. T., Quan J., Higgins S. G., et al. Predicting antibiotic resistance in gram-negative bacilli from resistance genes. Antimicrobial Agents and Chemotherapy . 2019;63(4):18. doi: 10.1128/aac.02462-18.024622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad M., Khan A. U. Global economic impact of antibiotic resistance: a review. Journal of Global Antimicrobial Resistance . 2019;19:313–316. doi: 10.1016/j.jgar.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 6.Almakki A., Jumas-Bilak E., Marchandin H., Licznar-Fajardo P. Antibiotic resistance in urban runoff. Science of the Total Environment . 2019;667:64–76. doi: 10.1016/j.scitotenv.2019.02.183. [DOI] [PubMed] [Google Scholar]
- 7.Barancheshme F., Munir M. Strategies to combat antibiotic resistance in the wastewater treatment plants. Frontiers in Microbiology . 2017;8:p. 2603. doi: 10.3389/fmicb.2017.02603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatima S., Liaqat F., Akbar A., et al. Virulent and multidrug resistant Klebsiella pneumoniae from clinical samples in Balochistan. International Wound Journal . 2021;18(4):510–518. doi: 10.1111/iwj.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akbar A., Sitara U., Ali I., Khan M. I., Phadungchob T., Anal A. K. Presence of Escherichia coli in poultry meat: a potential food safety threat. International Food Research Journal . 2014;21(3):941–945. [Google Scholar]
- 10.Wuijts S., van den Berg H. H. J. L., Miller J., et al. Towards a research agenda for water, sanitation and antimicrobial resistance. Journal of Water and Health . 2017;15(2):175–184. doi: 10.2166/wh.2017.124. [DOI] [PubMed] [Google Scholar]
- 11.Colquechagua Aliaga F., Sevilla Andrade C., Gonzales Escalante E. Enterobacterias productoras de betalactamasas de espectro extendido en muestras fecales en el Instituto Nacional de Salud del Niño, Perú. Revista Peruana de Medicina Experimental y Salud Pública . 2015;32(1):26–32. doi: 10.17843/rpmesp.2015.321.1571. [DOI] [PubMed] [Google Scholar]
- 12.Sahoo K. C., Tamhankar A. J., Sahoo S., Sahu P. S., Klintz S. R., Lundborg C. S. Geographical variation in antibiotic-resistant Escherichia coli isolates from stool, cow-dung and drinking water. International Journal of Environmental Research and Public Health . 2012;9(3):746–759. doi: 10.3390/ijerph9030746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talukdar P. K., Rahman M., Rahman M., et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One . 2013;8(4) doi: 10.1371/journal.pone.0061090.e61090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez L., Jashari T., Vasquez J., et al. Real-time PCR detection of Burkholderia cepacia in pharmaceutical products contaminated with low levels of bacterial contamination. PDA Journal of Pharmaceutical Science & Technology . 2018;72(1):73–80. doi: 10.5731/pdajpst.2017.007971. [DOI] [PubMed] [Google Scholar]
- 15.Jean B. P. Performance Standards for Antimicrobial Susceptibility Testing . Wayne, NJ, USA: Clinical and Laboratory Standards Institute Wayne; 2017. [Google Scholar]
- 16.Akbar A., Anal A. K. Isolation of Salmonella from ready-to-eat poultry meat and evaluation of its survival at low temperature, microwaving and simulated gastric fluids. Journal of Food Science & Technology . 2015;52(5):3051–3057. doi: 10.1007/s13197-014-1354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayne P. Clinical and Laboratory Standards Institute: performance standards for antimicrobial disk susceptibility tests; approved standard . Wayne, NJ, USA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 18.Randall L. P., Cooles S., Osborn M., Piddock L., Woodward M. J. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. Journal of Antimicrobial Chemotherapy . 2004;53(2):208–216. doi: 10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- 19.Toro C. S., Farfán M., Contreras I., et al. Genetic analysis of antibiotic-resistance determinants in multidrug-resistant Shigella strains isolated from Chilean children. Epidemiology and Infection . 2005;133(1):81–86. doi: 10.1017/s0950268804003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cebula T. A., Payne W. L., Feng P. Simultaneous identification of strains of Escherichia coli serotype O157: H7 and their shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. Journal of Clinical Microbiology . 1995;33(1):248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stacy-Phipps S., Mecca J. J., Weiss J. B. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. Journal of Clinical Microbiology . 1995;33(5):1054–1059. doi: 10.1128/jcm.33.5.1054-1059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. 2016. https://scholar.google.co.in/scholar?q=%22Tackling+Drug-Resistant+Infections+Globally:+Final+Report+and+Recommendations%22&hl=en&as_sdt=0&as_vis=1&oi=scholart .
- 23.WHO. No time to wait–securing the future from drug-resistant infections. Report to the Secretary General of the Nations . 2019 Geneva, Switzerland. [Google Scholar]
- 24.Shimpoh T., Hirata Y., Ihara S., et al. Prevalence of pks-positive Escherichia coli in Japanese patients with or without colorectal cancer. Gut Pathogens . 2017;9(1):35–38. doi: 10.1186/s13099-017-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momtaz H., Rahimi E., Moshkelani S. Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Veterinarni Medicina . 2012;57(4):193–197. doi: 10.17221/5916-vetmed. [DOI] [Google Scholar]
- 26.Nontongana N., Sibanda T., Ngwenya E., Okoh A. I. Prevalence and antibiogram profiling of Escherichia coli pathotypes isolated from the Kat River and the Fort Beaufort abstraction water. International Journal of Environmental Research and Public Health . 2014;11(8):8213–8227. doi: 10.3390/ijerph110808213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scaria J., Warnick L. D., Kaneene J. B., May K., Teng C.-H., Chang Y.-F. Comparison of phenotypic and genotypic antimicrobial profiles in Escherichia coli and Salmonella enterica from the same dairy cattle farms. Molecular and Cellular Probes . 2010;24(6):325–345. doi: 10.1016/j.mcp.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moges F., Endris M., Belyhun Y., Worku W. Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC Research Notes . 2014;7(1):215–216. doi: 10.1186/1756-0500-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odonkor S. T., Addo K. K. Prevalence of multidrug-resistant Escherichia coli isolated from drinking water sources. International Journal of Microbiology . 2018;2018:7. doi: 10.1155/2018/7204013.7204013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Agamy M. H., Shibl A. M., Hafez M. M., Al-Ahdal M. N., Memish Z. A., Khubnani H. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in Riyadh: emergence of CTX-M-15-producing E. coli ST131. Annals of Clinical Microbiology and Antimicrobials . 2014;13(1):4–7. doi: 10.1186/1476-0711-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidjabat H. E., Paterson D. L., Adams-Haduch J. M., et al. Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western Pennsylvania. Antimicrobial Agents and Chemotherapy . 2009;53(11):4733–4739. doi: 10.1128/aac.00533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyimo B., Buza J., Subbiah M., Smith W., Call D. R. Comparison of antibiotic resistant Escherichia coli obtained from drinking water sources in northern Tanzania: a cross-sectional study. BMC Microbiology . 2016;16(1):254–310. doi: 10.1186/s12866-016-0870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aleem M., Azeem A. R., Rahmatullah S., Vohra S., Nasir S., Andleeb S. Prevalence of bacteria and antimicrobial resistance genes in hospital water and surfaces. Cureus . 2021;13(10):p. e18738. doi: 10.7759/cureus.18738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibekwe A. M., Murinda S. E., Graves A. K. Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. PLoS One . 2011;6(6) doi: 10.1371/journal.pone.0020819.e20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grevskott D. H., Ghavidel F. Z., Svanevik C. S., Marathe N. P. Resistance profiles and diversity of β-lactamases in Escherichia coli strains isolated from city-scale sewage surveillance in Bergen, Norway mimic clinical prevalence. Ecotoxicology and Environmental Safety . 2021;226 doi: 10.1016/j.ecoenv.2021.112788.112788 [DOI] [PubMed] [Google Scholar]
- 36.Kirtikliene T., Mierauskaitė A., Razmienė I., Kuisiene N. Genetic characterization of multidrug-resistant E. coli isolates from bloodstream infections in Lithuania. Microorganisms . 2022;10(2):p. 449. doi: 10.3390/microorganisms10020449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derakhshan S., Farhadifar F., Roshani D., Ahmadi A., Haghi F. Study on the presence of resistant diarrheagenic pathotypes in Escherichia coli isolated from patients with urinary tract infection. Gastroenterology and Hepatology from Bed to Bench . 2019;12(4):348–357. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Major part of the data is presented in the manuscript, and the remaining data will be made available on reasonable request.


