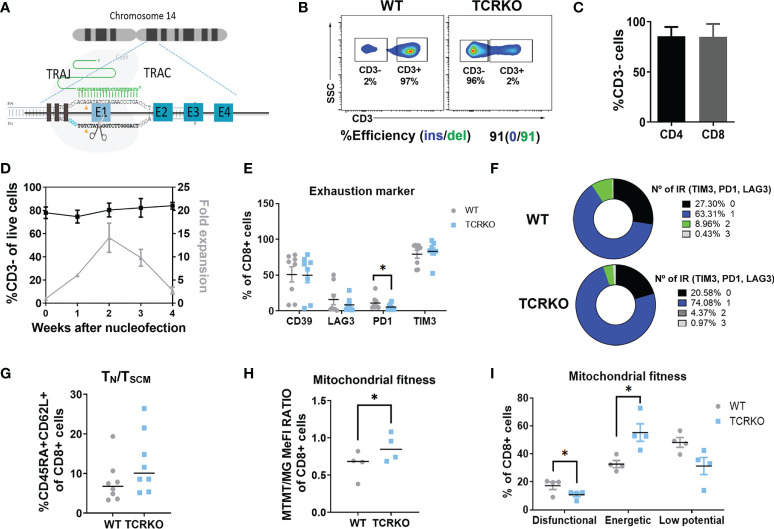
Figure 1.
Generation of TCR negative T cells by Cas9/gRNATCRA RNP electroporation. (A) Diagram of CRISPR/Cas9 system composed by a specific gRNA against exon 1 (RNPTRAC) of the TRAC locus and Cas9 recombinant protein. (B) Efficiency of TCR disruption in primary T cells at day 3 post-nucleofection. CD3 expression by flow cytometry (top plots) and ICE analysis of PCR product (bottom) are shown. (C) Graph showing % of CD3- in CD4+ and CD8+ T cell populations (4 independent donors, n=8). (D) Graph showing % of CD3- of edited T cells (left Y axis) and expansion over time (right Y axis) (4 independent donors, n=4). (E) Percentage of CD39+, LAG3+, PD1+ and TIM3+ of CD8+ mock electroporated (WT) and TCRKO T cells (F) Spice graphs showing the percentage of expression of 0(black), 1(blue), 2(green) and 3(gray) exhaustion markers/inhibitory receptors (IR) (TIM3, PD1 and LAG3) in CD8+ WT (left) and TCRKO (right) T cells (G) Percentage of TN/TSCM CD8+ T cells, determined as CD45RA+CD62L+ (7 independent donors, n=8). (H) Quantification of flow cytometry data at day 10 post activation showing the ratio of mitochondrial membrane potential (MTRM) to mitochondrial mass (MG) in TCRKO and mock electroporated CD8+ T cells and (I) Percentage of dysfunctional(MG+TMRM-), energetic (MG+TMRM+) and low potential mitochondria(MG-TMRM-) in TCRKO and mock electroporated CD8+ T cells (WT) (4 independent donors, n=4).Statistics are based on paired, two-tailed Student´s t-test (H, I), *p<0.05. Graphs show mean ± SEM.