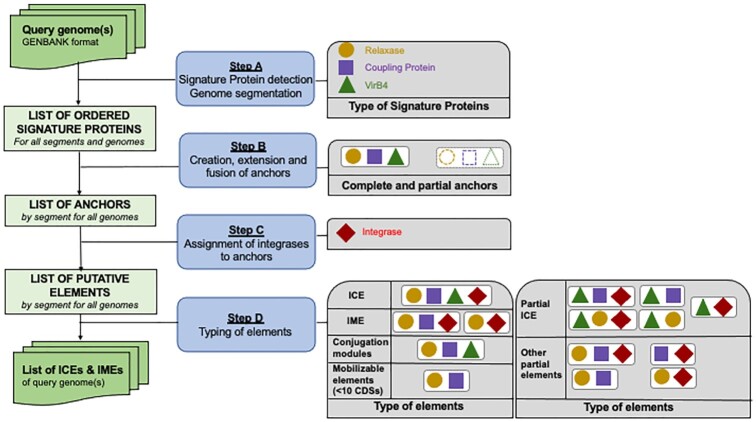
Figure 1.
The four main steps of the ICEscreen workflow. The input query genome(s) must be annotated genomes in GenBank format. Step A: The first step consists in detecting three types of Signature Proteins (SPs), which are relaxases, coupling proteins and VirB4. The resulting ordered list of SPs per genome is then segmented into groups of SPs where the distance between two subsequent SPs is less than 100. Step B: The second step consists in searching for all potential conjugation modules by ‘creating, extending and fusing’ anchors. An anchor is created when a relaxase, a coupling protein or a VirB4 is detected. Then, the anchor is extended left and right to detect group of SPs (excluding Integrases) corresponding to putative elements. When all potential anchors have been created, the algorithm evaluates the possibility of combining anchors in order to resolve the cases of complex structures (for instance nested elements). Step C: The integrase is then searched on both sides of each anchor. Step D: The next step consists in typing each element according to its SP content and size as described in the two panels on the right.