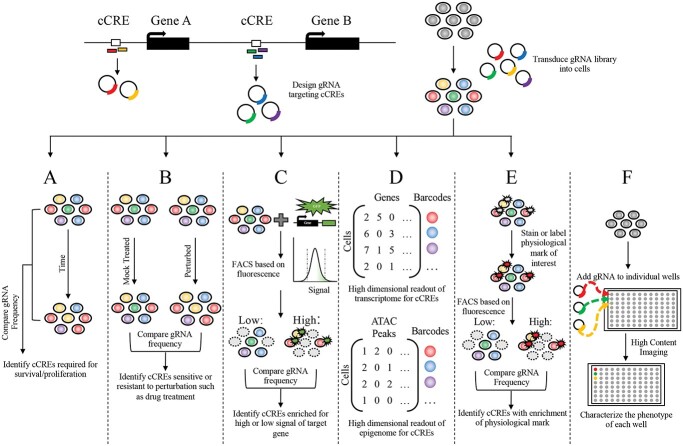
Figure 2.
Strategies of CRISPR screens. Experiment overview for high-throughput CRISPR screening, where gRNAs designed to target cCREs are cloned and then delivered into the cell type of interest containing CRISPR machinery. After delivery, the transduce cells containing the pooled gRNAs can be characterized by several different techniques with varying readouts. (A) Comparing the distribution of pooled gRNAs between two time points can identify loci essential for proliferation and survivability. (B) Assessing the relative depletion or enrichment of gRNAs between a perturbed condition such as drug treatment compared with a mock treatment at the same time point or the time point before treatment, connects loci that confer sensitivity of resistance to the perturbation. (C) Quantifying the expression changes induced by cCREs of an endogenous gene tagged with GFP allows FACS to separate cells with high or low levels of fluorescence/gene expression and identify loci that regulate the expression of the tagged gene. (D) Conducting scRNA-seq or scATAC-seq identifies high-dimensional transcriptional or epigenomic phenotypes associated with perturbation of cCREs. (E) Separating pooled cells, for example by FACS, with a labeled physiological biomarker such as protein levels, allows for the gRNA distribution of cells with and without the marker to be compared with identify loci associated with a biological process. (F) Complex phenotypes that cannot be measured in pooled screens can be investigated in arrayed screens where gRNAs are introduced and characterized in individual wells.