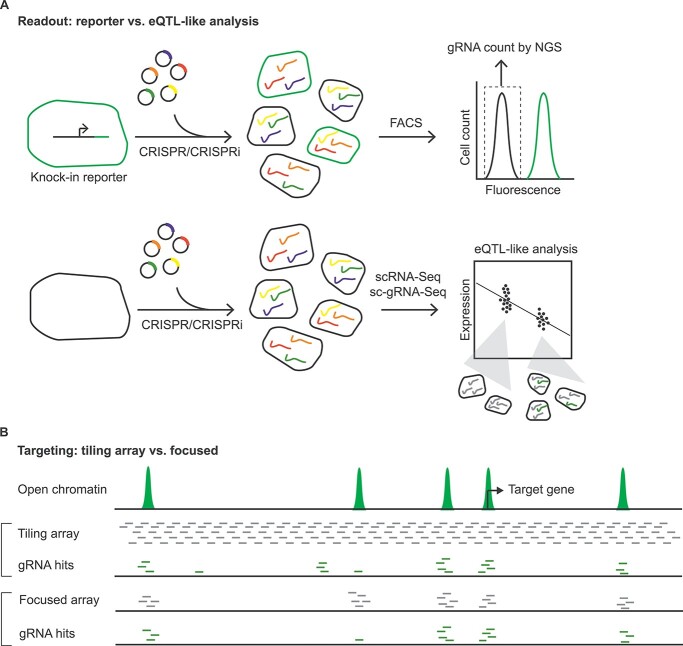
Figure 2.
Common screening strategies for the noncoding regulatory landscape with CRISPR-based tools. (A) In the first strategy (upper panel), a cellular knock-in reporter line is constructed to monitor endogenous expression of the target gene. Following CRISPRi or CRISPR perturbation, cells with low reporter expression are identified by FACS, followed by gRNA quantification to identify enrichment within functional relevant sites. This type of screening is inexpensive and well-powered, but limited to one target gene. In the second strategy (lower panel), single-cell RNA-seq (scRNA-seq) is performed along with single-cell gRNA sequencing (sc-gRNA-seq). Cells expressing each unique gRNA are grouped together and compared with the remaining cells to identify differentially expressed genes; this analytic framework is similar to eQTL analysis and offers genome-wide coverage, but requires much greater sequencing depth and expense. (B) Screening can be performed in either a tiling (upper) or focused (lower) fashion. The tiling array is unbiased but limited to a restricted number of loci, while the focused array is biased toward known epigenomic or enhancer annotations but enables surveillance of many distinct loci simultaneously.