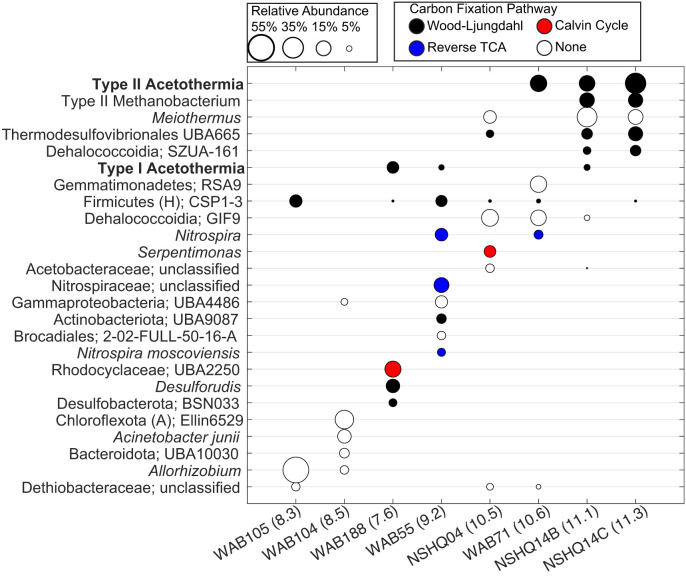
Fig. 3.
Putative autotrophs in subsurface fracture water communities from the SO. Each column shows the relative abundances of reconstructed MAGs (>5% relative abundance) within each well water community, with the corresponding taxonomic classification of MAGs given on the left. The circles represent individual MAGs and are scaled based on the relative abundance of that MAG among others within that community (based on the legend at the top left of the plot). The circles are colored according to the autotrophic carbon fixation pathway that is inferred to be encoded within the MAG, as based on the legend at the top right of the plot. Well names are followed by the pH of the sampled fracture waters in parentheses. Taxonomic classifications are given at the lowest characterized taxonomic designation, followed by either the specific uncultured group it belongs to (alphanumeric designations) or are otherwise followed by “unclassified” if the MAG is not related to previously characterized genomes within the Genome Toolkit Database. The type II Methanobacterium is the same as described in Fones et al. (27), and, while it encodes the WL pathway, it is dependent on formate as an electron donor and carbon source. The 2015 and 2020 metagenomes were not used for abundance calculations, given their relatively low sequencing depth and limited sampling scope, respectively.