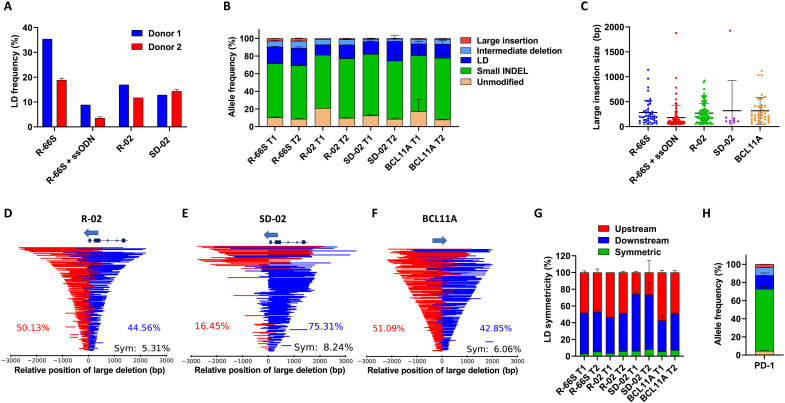
Fig. 3. Unintended large gene modifications are common for CRISPR gRNAs.
(A) Comparison of the LD rates in SCD HSPCs from two SCD patients (Donor #1 and Donor #2) analyzed by SMRT-seq for R-66S RNP with and without corrective ssODN, R-02 RNP, and SD-02 RNP. (B) SMRT-seq with UMI-based quantification of LD, intermediate deletion, large insertion, and small INDEL. gDNAs from SCD HSPCs edited by R-66S, R-02, SD-02, and BCL11A RNPs were collected on day 4 after delivery (T1) and from cells after 14 days of erythroid differentiation (on day 17 after delivery) (T2). High frequencies of diverse LDs of up to 4 kb and insertions of up to 1.9 kb were found at all loci tested. After 2 weeks of erythroid differentiation of SCD HSPCs, the LDs persisted and their rates increased. (C) Large insertion size distribution (50 bp to 1.9 kb) in RNP-treated samples from Donor #2. (D) LD profile (location and size) at HBB in the R-02 RNP-treated sample. (E) LD profile at HBG1 in the SD-02 RNP-treated sample. (F) LD profile at BCL11A in the BCL11A RNP-treated sample. In (D) to (F), schematics of HBB and HBG1 genes are shown to scale; the schematic of BCL11A gene is not shown; arrows show the 5′ to 3′ orientation of the gRNAs. (G) Frequencies of LD positions relative to the Cas9 cut site (upstream, downstream, or symmetric) for four RNP-treated samples at T1 and T2. (H) SMRT-seq with UMI quantification of allele frequency in PD-1 RNP-treated primary human T cells. The color code is the same as in (B).