Figure 5.
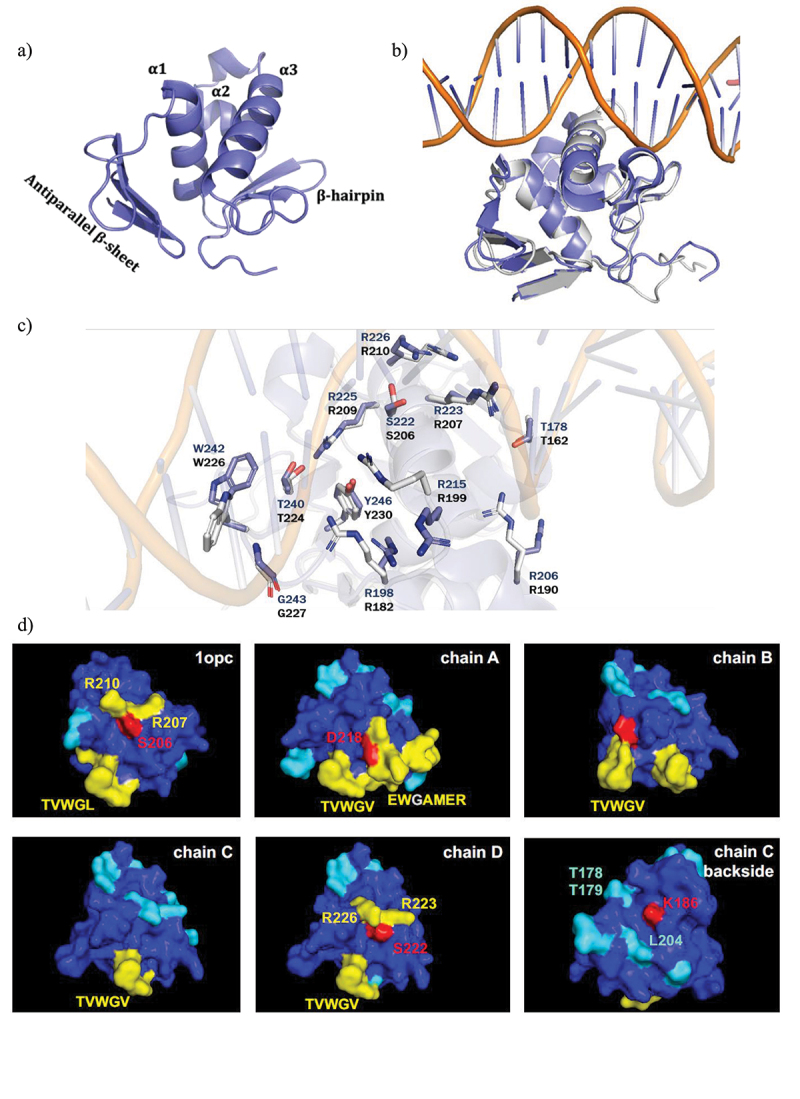
Structure of the DNA-binding domain of A. baumannii OmpR and computational hotspots analysis.
A) Overview of the crystal structure of the DNA-binding domain (DBD) of A. baumannii OmpR. B) Superposed crystal structure of OmpR DBD from A. baumannii (blue) on DNA bound structure of OmpR from E. coli (grey). C) Amino acids (blue colored for OmpRABA and grey colored for OmpRECO) that are involved in making H-bond and salt bridges with DNA are shown. D) The 3D structures of OmpRECO (1opc) and OmpRABA DBD (chain A, B, C and D) are represented and colored based on temperature scale from blue (low ligand interaction probability) to red (high ligand interaction probability). Individual residues with high predicted ligand-binding potential are highlighted. The interaction site is around the conserved residues TVWG(V/L). Hotspot 1 in OmpRABA-DBD chain A contains D218, as anchor residue for ligand binding. Hotspot 2 in OmpRABA-DBD chain D contains residues S222, R223 and R226 (S206, R207 and R210 in OmpRECO DBD). Hotspot 3 in OmpRABA-DBD chain C contains residues K186, L204, and T178/T179.
