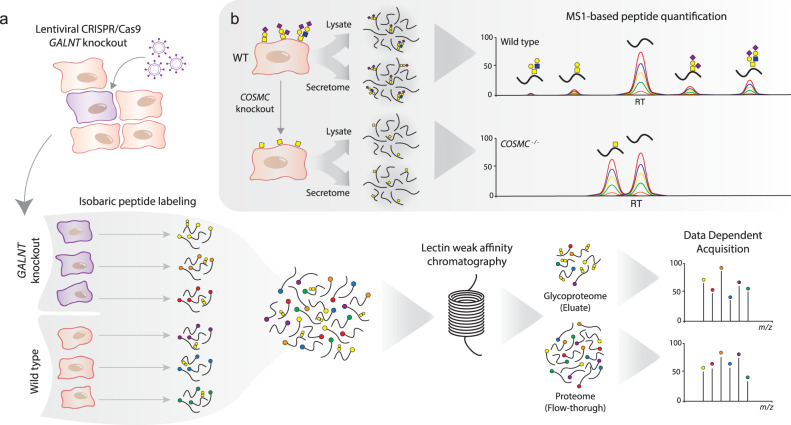
Fig. 1. Scheme of the applied workflow.
a GALNT knockouts in N/TERT-1 cell lines were generated using lentiviral transduction to deliver the CRISPR/Cas9 machinery. Total cell lysates were collected from three technical wild-type replicates and three biological replicate knockout clones, and extracted proteins were reduced, alkylated, and digested with trypsin. Purified peptides were labeled using tandem mass tags (TMT), mixed, and treated with neuraminidase prior to jacalin LWAC enrichment of glycopeptides. Finally, the enriched sample was separated into eight high pH fractions and simultaneous discovery and reporter quantification were performed using LC-MS/MS. b For the assessment of site occupancy, both total cell lysates and secreted material were obtained from three technical replicates of wild type and COSMC knockout cells. Both sample types were reduced, alkylated, and digested with trypsin followed by N-glycan removal using PNGase F. Each sample was then separated into four high pH fractions and analyzed using LC-MS/MS, where relative occupancies were calculated based on MS1 peak intensity from glycosylated peptides and their non-glycosylated equivalents.