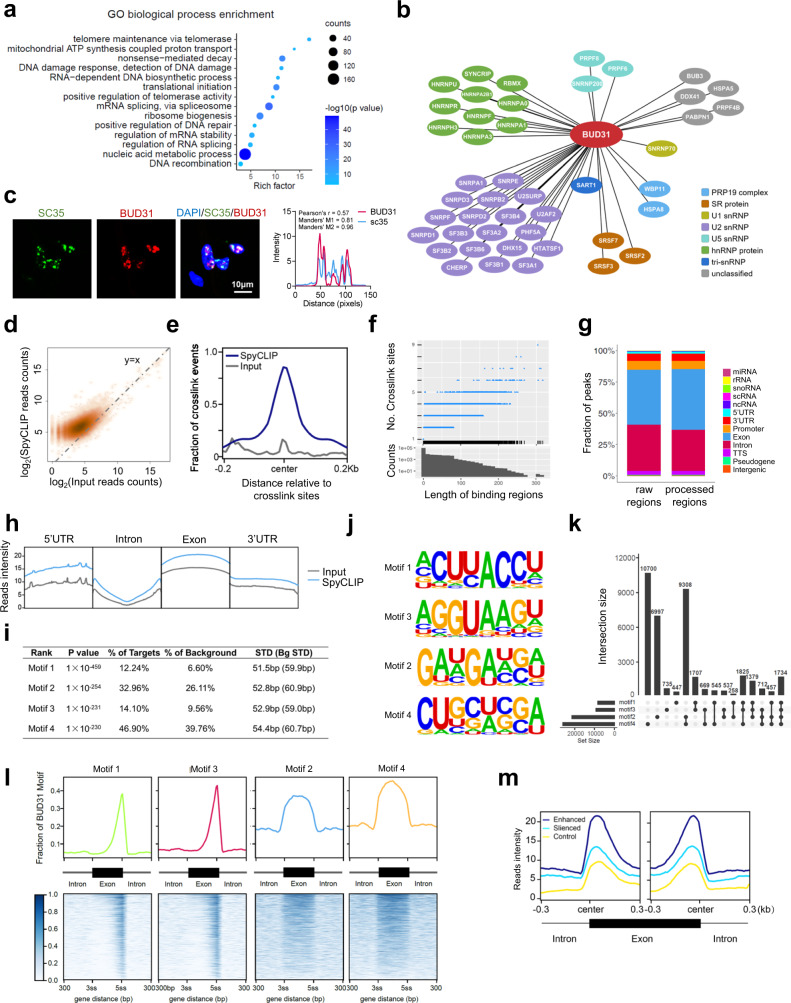
Fig. 4. Identification of the genome-wide BUD31-binding sites on RNA.
a GO biological process enrichment analysis of BUD31-interacting proteins in HEYA8 cells using immunoprecipitation coupled to mass spectrometry. b The correlation network between BUD31 and splicing factors was constructed in Cytoscape. Proteins belonging to the spliceosome were classified into PRP19 complex, SR protein, U1 snRNP, U2 snRNP, U5 snRNP, hnRNP protein, tri-snRNP, and others. c Immunofluorescence assays showed the co-localization of BUD31 (red) with splicing factor SC35 (green) in punctate nuclear speckles. The quantification and analysis of co-localization was performed with Coloc 2 and Plot Profile (Pearson’s r = 0.57, Manders’ M1 = 0.81, Manders’ M2 = 0.96). d High-density scatter plot of the SpyCLIP and Input reads counts aligned to the BUD31-binding regions. e Position of the SpyCLIP crosslinking regions relative to the crosslinking sites identified by PURECLIP. The SpyCLIP (deep blue) and input (gray) signals are shown around the crosslink sites. f Length distribution of the BUD31-binding regions and the crosslinking sites included in the corresponding binding regions. g Distribution of SpyCLIP crosslinking regions annotated by HOMER on genome elements. Processed regions were longer than 3 nucleotides. h SpyCLIP read distribution compared with input on the genome elements, including 5′ UTR, intron, exon, and 3′ UTR. i De novo motif analysis of BUD31 SpyCLIP clusters and statistical results of the top-four BUD31-binding motifs ranked by HOMER calculated p value. j Enriched sequence elements of the top-four BUD31-binding motifs. k Upset plot of the distribution of motifs 1–4 in the BUD31-binding regions. l BUD31-binding motif distribution in the exon region and the 300 bp flanking the 3ss or 5ss intron-exon junction site. The exon region was scaled such that the length was equal to 300 bp to normalize different exon lengths. m SpyCLIP reads intensity distributed around the alternative exons. Enhanced exons (deep blue) were included, and silenced exons (blue) were excluded after silencing BUD31. Randomly chosen exons were used as controls (yellow).