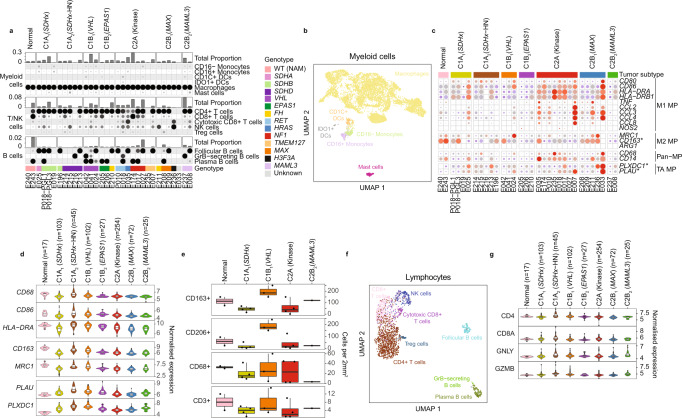
Fig. 4. Analysis of leukocytes isolated from PCPG and NAM tissues by snRNA-seq.
a Supervised classification of immune cell fractions in PCPG. Proportions of each major immune cell types within each sample (top panel) and the functional subsets that make up each major cell type (bottom panels). b UMAP re-clustering of myeloid cells from all snRNA-seq samples. Nuclei are colored according to their classified functional cell subsets (nCells = 7977). c snRNA-seq expression of macrophage (MP) marker genes in macrophages detected on PCPG and NAM tissues. (TA: Tumor-associated) d Enumeration of intra-tumoral leucocytes detected by immunohistochemistry staining for canonical macrophage markers (CD163, CD206, CD68) and a T cell marker (CD3) (nSamples = 645, the lower and upper hinges of each boxplot correspond to the first and third quartiles, respectively, and the median value is marked. The whiskers extend to the largest and smallest value not greater than 1.5 times the interquartile range above or below the upper and lower hinges, respectively. Values beyond the whisker extents are deemed outliers and are plotted individually). e Bulk-tissue expression of DE expressed macrophage genes contrasting tumor and NAM macrophages (nSamples = 14, box plots to be interpreted as per panel d). f UMAP reclustering of T/B cells from all snRNA-seq samples (nCells = 2140). g Bulk-tissue gene-expression of T/NK -cell markers across subtypes. DCs dendritic cells, NK cells natural killer cells. WT (NAM) wild type normal adrenal medulla (nSamples = 645, box plots to be interpreted as per panel d).