Figure 11.
Validation of DA neuron Shh deletion
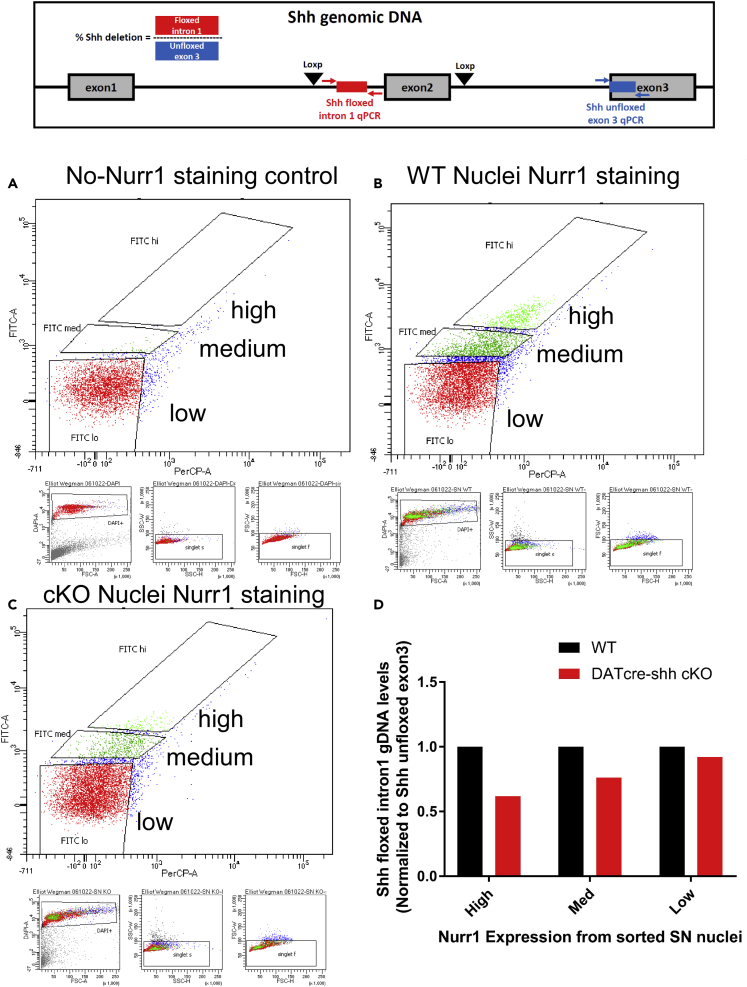
DA neuron nuclei were enriched by Nurr1 immunoreactivity fluorescence-activated nuclear sorting (FANS) from the SN of two WT and three DATcre-shh cKO mice. Nuclei were sorted into three categories: high expression (1.5–2.5k nuclei), medium expression (17k-18k nuclei), and low expression (150–200k nuclei) of Nurr1. Depicted are the gates for the high, medium, and low expression populations in (A) a control sample not stained for Nurr1, (B) the pooled WT sample, and (C) the pooled DATcre-shh cKO sample. Genomic DNA from the sorted nuclei was harvested and qPCR performed for the floxed Shh intron 1 and unfloxed exon 3 to confirm the deletion of the floxed site in DATcre-shh cKO DA nuclei. Shh floxed intron 1 gDNA levels were normalized to unfloxed exon 3 gDNA levels in each sample. The data show a ∼30% decrease in floxed Shh intron 1 gDNA harvested from Nurr1+ nuclei in DATcre-shh KO mice compared to WT mice (D) while levels of Shh floxed intron 1 do not change in Nurr1-negative nuclei. These results are in line with a previous study that used this method to enrich for DA nuclei and observed ∼25% abundance of DA neuron nuclei in the sorted Nurr1+ population (Kamath et al., 2022).