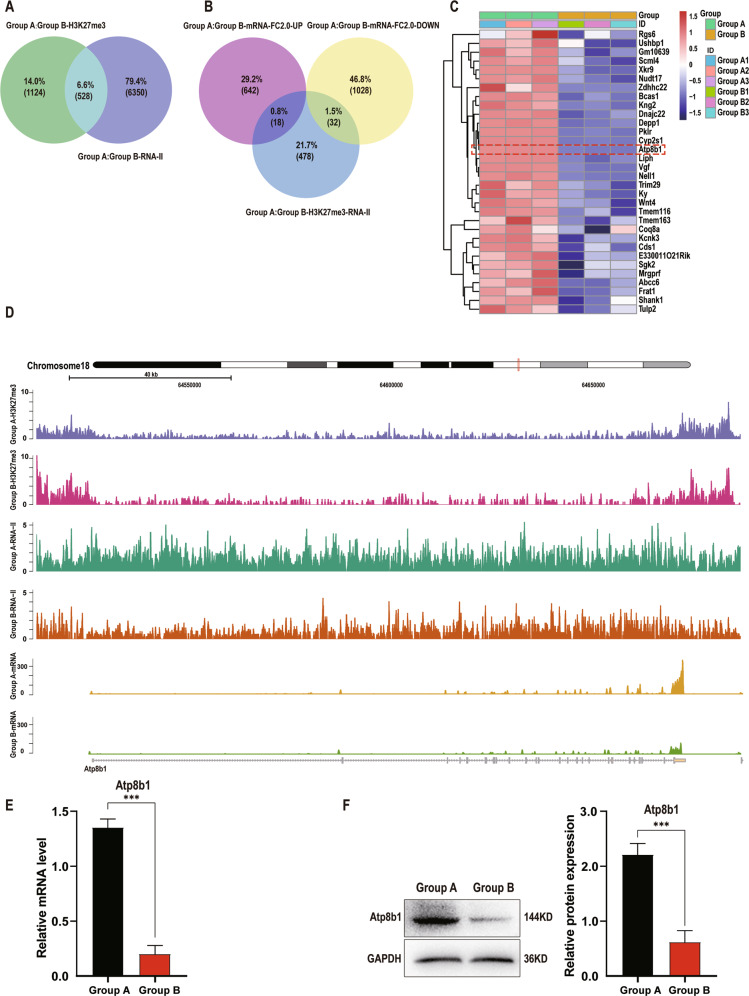
Fig. 2. Atp8b1 is downregulated in CP tissues from PRSS1Tg mice.
H3K27me3 ChIP-seq, RNA pol II ChIP-seq and mRNA-seq analyses of pancreatic tissues from Group A (treated with caerulein for 1 week) and Group B (treated with caerulein for 4 weeks) were applied to identify genes that potentially regulate phospholipid metabolism. A Venn diagram analysis of the overlap between the H3K27me3 ChIP-seq and RNA pol II ChIP-seq. B Venn diagram analysis of the overlap between the above 528 genes and differentially expressed mRNAs in Group A and Group B separately. C Heatmap of the 32 downregulated genes. D Signal tracks showing the binding and co-occupancy of H3K27me3 and RNA pol II at the Atp8b1 gene locus and mRNA-seq signals for genes in pancreatic acinar cells in Groups A and B. Purple, Group A-H3K27me3; Red, Group B-H3K27me3, Green, Group A- RNA pol II; Orange, Group B- RNA pol II; Yellow, Group A-mRNA; Cyan-blue, Group B- mRNA. E The total Atp8b1 mRNA and F protein expression levels in pancreatic tissue in Group A and B were measured by quantitative RT-PCR and western blotting, respectively. The results were measured by densitometry and expressed by expression relative to GAPDH as the reference protein. The data are presented as the means ± SDs; n = 3 biological replicates. ***p ≤ 0.001.