Figure 6.
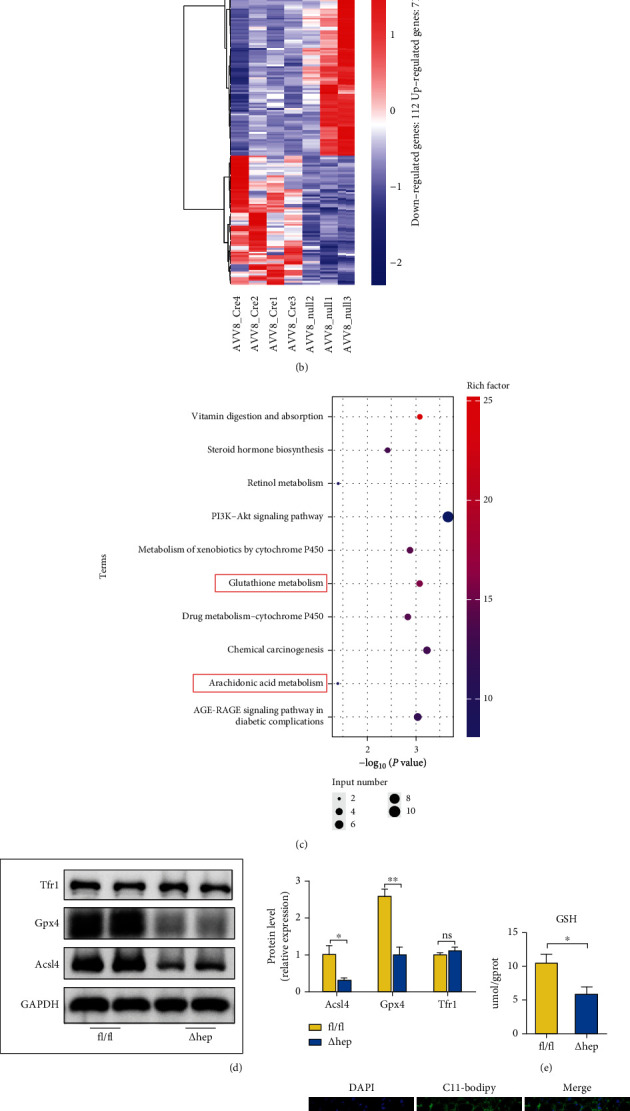
Hepatocyte-specific knockout of Per2 might alleviate NASH by inhibiting ferroptosis. (a) Volcano plot showing upregulated and downregulated genes in Per2△hep and Per2fl/fl mice. (b) Heatmap showing the differentially expressed genes (DEGs) between Per2△hep and Per2fl/fl mice; blue indicates low expression and red indicates high expression. (c) The DEGs between Per2△hep and Per2fl/fl mice were identified with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Rich factor (%) was the ratio of the number of DEGs annotated in a pathway (as indicated in the y-axis) to the number of all genes annotated in the pathway. (d) Acsl4, Gpx4, and Tfr1 protein levels were measured by western blotting, and GAPDH was used as a control. (e) Liver GSH concentrations were quantified by a commercial kit. (f) Electron microscopic evaluation of the morphologic changes in mitochondria; the white arrow indicates lipid droplets and the black arrow indicates mitochondria (original magnification, 5000x; scale bar, 1 μm) (Per2fl/fl, n = 2; Per2△hep, n = 3). (g) Fluorescence staining represents lipid peroxidation (LPO) levels in the livers of Per2△hep and Per2fl/fl mice. The nuclei were labeled with DAPI (left), and LPO (middle) was labeled by C11-Bodipy (original magnification, 400x; scale bar, 50 μm). (h) Liver MDA concentrations were quantified with a commercial kit. (i) Serum and liver iron concentrations were determined using a commercial kit to compare the iron-overload levels. Both Per2△hep and Per2fl/fl mice were fed with HFHFD to induce NASH. Data are presented as mean ± SEM. ∗P < 0.05 and ∗∗P < 0.01; ns: not significant (also refer to Figure S2).
