Figure 1.
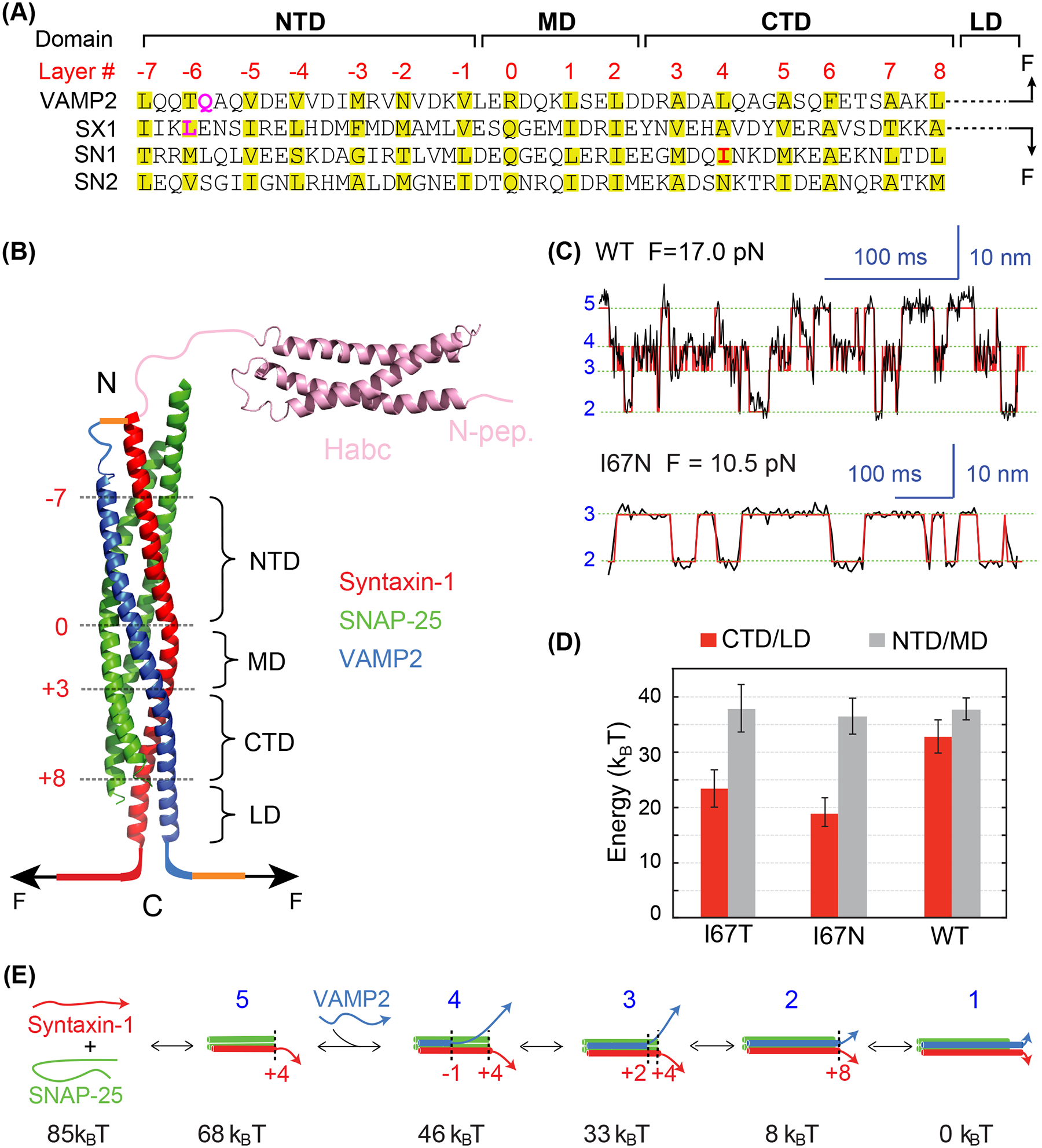
Energetics of stepwise SNARE zippering revealed by optical tweezers. (A) Amino acid sequences of SNARE motifs of synaptic SNAREs VAMP2, syntaxin-1A (SX1), and SNAP-25B (SN1 and SN2), and their associated folding domains and layer numbers. The folding domains include the N-terminal domain (NTD), the middle domain (MD), the C-terminal domain (CTD), and the linker domain (LD). The residues in VAMP2 Q36 and syntaxin-1 L205 in the red rectangle are mutated to cysteine and crosslinked to facilitate detection of reversible SNARE assembly in some single-molecule manipulation experiments (see results in C-E). (B) Crystal structure of the synaptic SNARE complex and its associated folding domains and layer numbers (red). The yellow lines indicate the disulfide bonds used to crosslink syntaxin and VAMP2 at their N-termini (top) and VAMP2 to the DNA handle (bottom, as in Figure 3(A)). The N-terminal SNARE crosslinking site varies in different experiments. (C) Time-dependent extension of the wild-type (top) or mutant (bottom) SNARE complex showing their reversible and stepwise zippering at constant force. The mutant contains a disease mutation at SNAP-25B I67N (highlighted red in A). The red lines represent idealized transitions derived from hidden-Markov modelling (Zhang et al. 2016). The green lines indicate average positions of the states with blue numbers shown in E. (D) Folding energy of different domains of the wild-type and mutant SNARE complexes. The disease mutations SNAP-25B I67N and I67T reduce the total folding energy of CTD and LD (CTD/LD), but not that of the NTD and MD (NTD/MD). (E) Diagrams of the different SNARE folding/assembly states and their energy relative to the fully assembled state.
