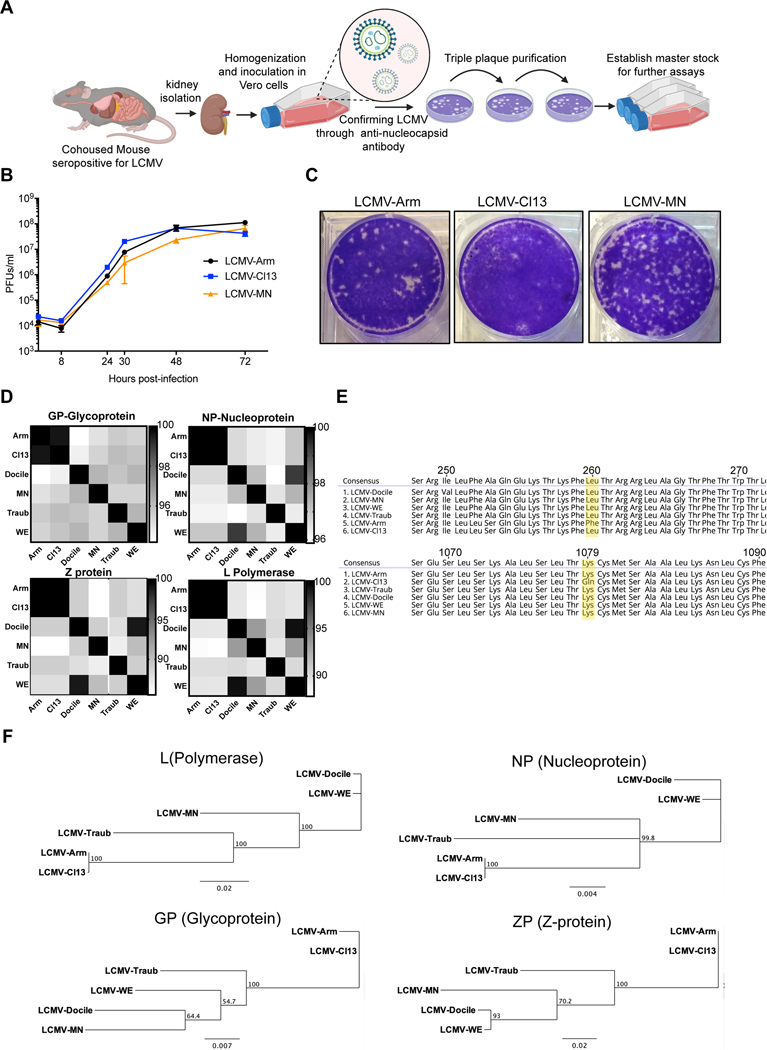
Figure 1. Isolation, in vitro growth and genomic characterization of LCMV-MN.
(A) Schematic of the process depicting isolation of a novel LCMV like virus from kidney homogenates of C57Bl/6 mice that were cohoused with petstore mice. The initial virus stock was confirmed via positive reactivity to LCMV-N monoclonal antibody (VL-4) and was triple plaque purified to establish final stock. (B) Comparative growth kinetics among LCMV-Armstrong, -Cl13 and -MN showing viral output from BHK-21 cells infected at an MOI of 0.1. Supernatant was collected at times indicated and titered on Vero cells using plaque assay. (C) Plaque morphology of the indicated viruses on Vero cells. (D) Amino acid percentage homology among various established LCMV strains and LCMV-MN across the four viral proteins (indicated at the top of the respective checkered box). The scale for each of the protein comparisons is on the right of the panel and represents percent identity. (E) Comparison of amino acid sequences across select stretched of glycoprotein (top) and polymerase protein (bottom) with a particular emphasis on conservation of GP260 and LP1079 residues among LCMV-MN and indicated strains. (F) Phylogenetic tree analysis output of the 4 proteins of LCMV-MN and other LCMV strains based on neighbor-joining method. Data in B are representative of two separate experiments with 3 replicate each per time point. Plaque morphology in ‘D’ is representative of 3 independent repeats. Bars indicate mean ± SEM. Figure in A created with Biorender (Biorender.com).