Figure 1. DGY-06–177 induced degradation of BRD4.
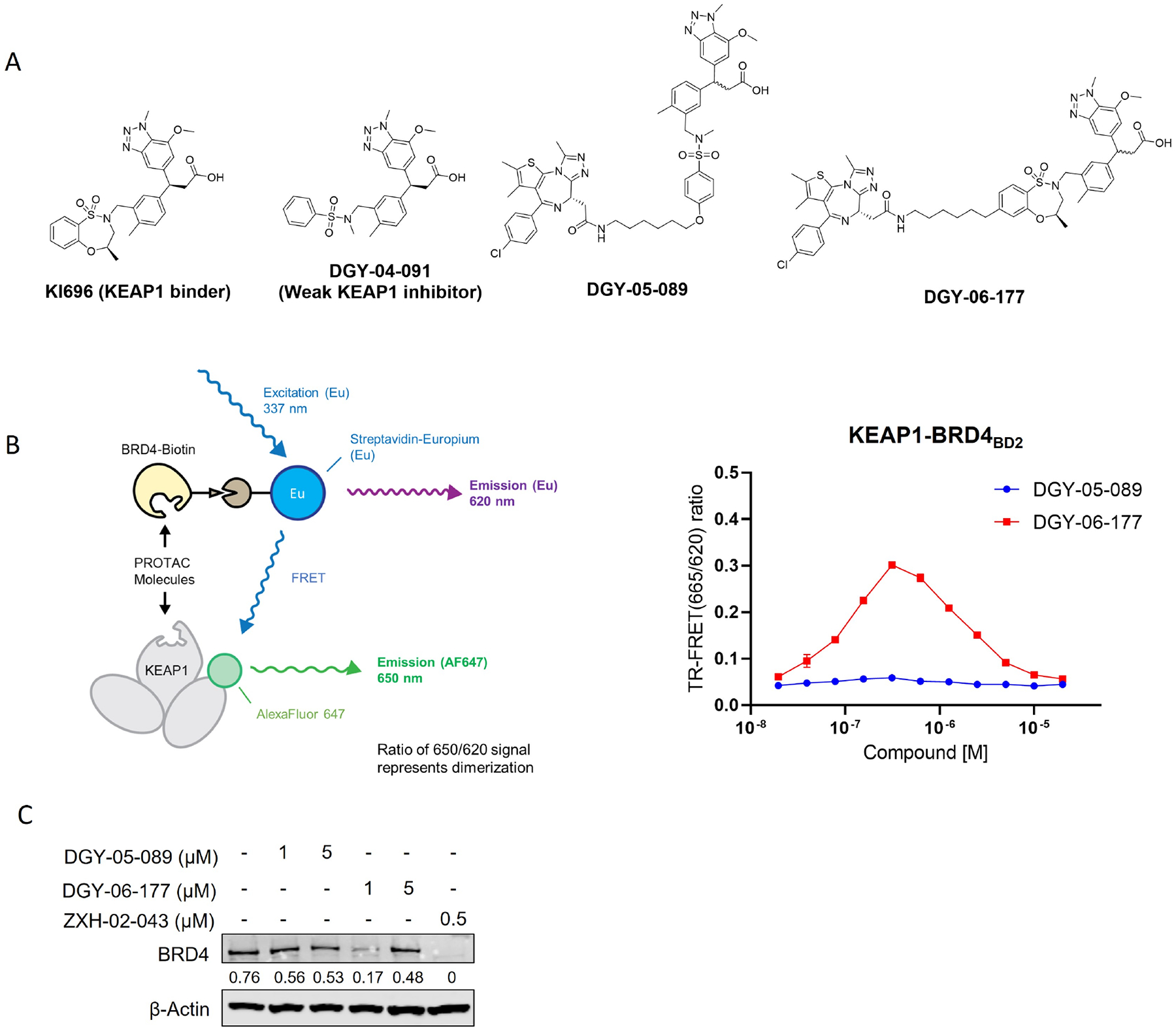
A. Chemical structures of KI-696, DGY-04–091, DGY-05–089 and DGY-06–177. B. KEAP1-BRD4BD2 dimerization TR-FRET assay. Titration of DGY-06–177 or DGY-05–089 to Alexa Fluor™ 647-KEAP1, Eu-streptavidin and biotinylated BRD4BD2. Data are presented as mean ± SD with at least three technical replicates. Representative figure from two independent experiments is shown. C. Immunoblot assessment of MM1.S cells treated with either DMSO or indicated compounds for 24 h. Representative blots from two independent experiments are shown. The relative intensity of each band (BRD4 normalized to β-actin) is shown under each band. See also Figure S1 and Table S1.
