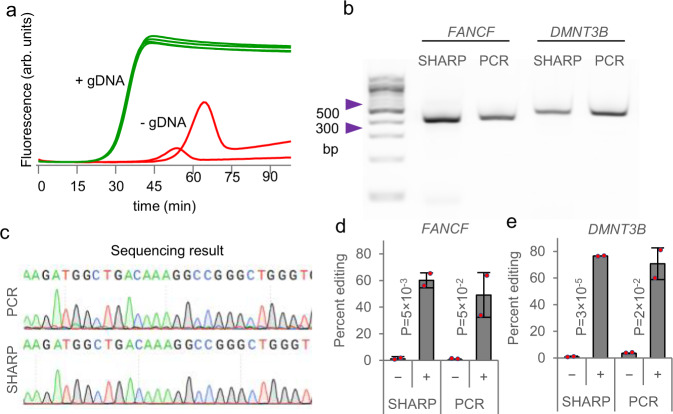
Fig. 4. SHARP applied to genomic DNA.
a The SHARP reaction containing gDNA template (+gDNA) or without template (–gDNA), along with primers intended to amplify a 474 bp region near the DMNT3B gene, was incubated at 60 °C. The extent of amplification was quantified by measuring DNA fluorescence using the EvaGreen dye as a function of time. b Agarose gel electrophoresis of genomic amplification product using SHARP versus PCR, amplifying a 410 bp region near FANCF or a 474 bp region near DMNT3B. c Sanger sequencing of genomic amplification product near DMNT3B using SHARP versus PCR. d-e Quantification of genome editing insertion-deletion mutations (indels) using SHARP versus PCR, near FANCF and DMNT3B as panel b. Signs ‘–‘ and ‘+’ correspond to gDNA from cells without or with Cas9/gRNA, respectively. Cas9/gRNA has been introduced to two biologically independent cell cultures. Data are presented as mean values ± standard deviation. Based on P-values obtained with Student’s t-test, SHARP quantified that the percent editing in cells with introduced Cas9/gRNA (+) is statistically significant relative to unedited (–) cells with the confidence level of 95%. Source data are provided as a Source Data file.