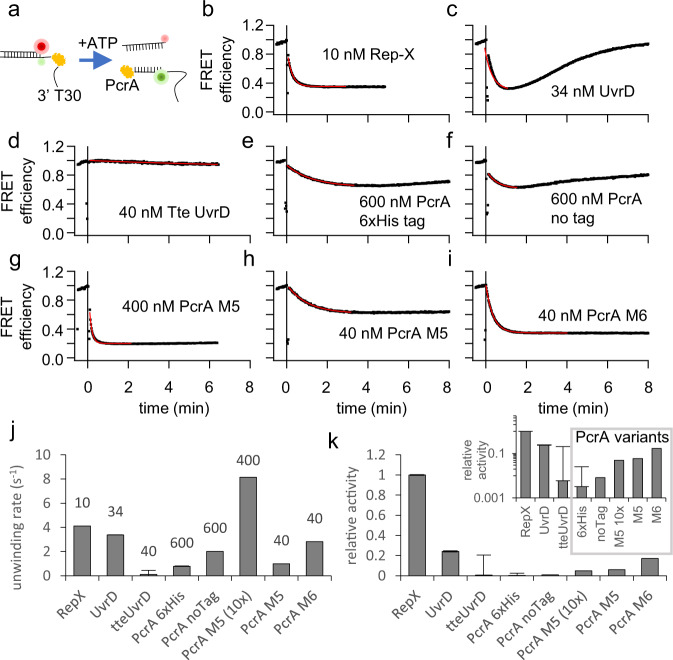
Fig. 6. Bulk unwinding activity of different DNA helicases at 37 °C.
Each helicase is at different concentration, while all other conditions are fixed. a Unwinding reaction of FRET-pair labelled DNA construct. b Rep-X superhelicase shows the highest unwinding activity even at a low concentration of 10 nM. c E. coli UvrD helicase at 34 nM shows high initial unwinding activity, but the reaction terminates quickly resulting in the DNA reannealing likely due to ATP depletion. d Thermostable Tte UvrD helicase at 40 nM shows very low unwinding activity at 37 °C. e Wild-type PcrA expressed with 6xHis tag shows low unwinding activity at 600 nM. f Wild-type PcrA without 6xHis tag shows low unwinding activity at 600 nM. g PcrA M5 mutant at 400 nM shows high unwinding activity. h PcrA M5 mutant at 40 nM. i PcrA M6 shows higher unwinding activity than M5 even at 40 nM concentration. SHARP uses nearly 40 nM of PcrA M6. j Unwinding rates obtained from the fit to FRET efficiency in b to i. k Unwinding activity relative to Rep-X is obtained by dividing unwinding rates in j by the concentration and normalizing. Inset shows relative activity on log10 axis. Data in j and k are presented as mean values ± standard error obtained from the fit. Source data and fitting parameters are provided as a Source Data file.