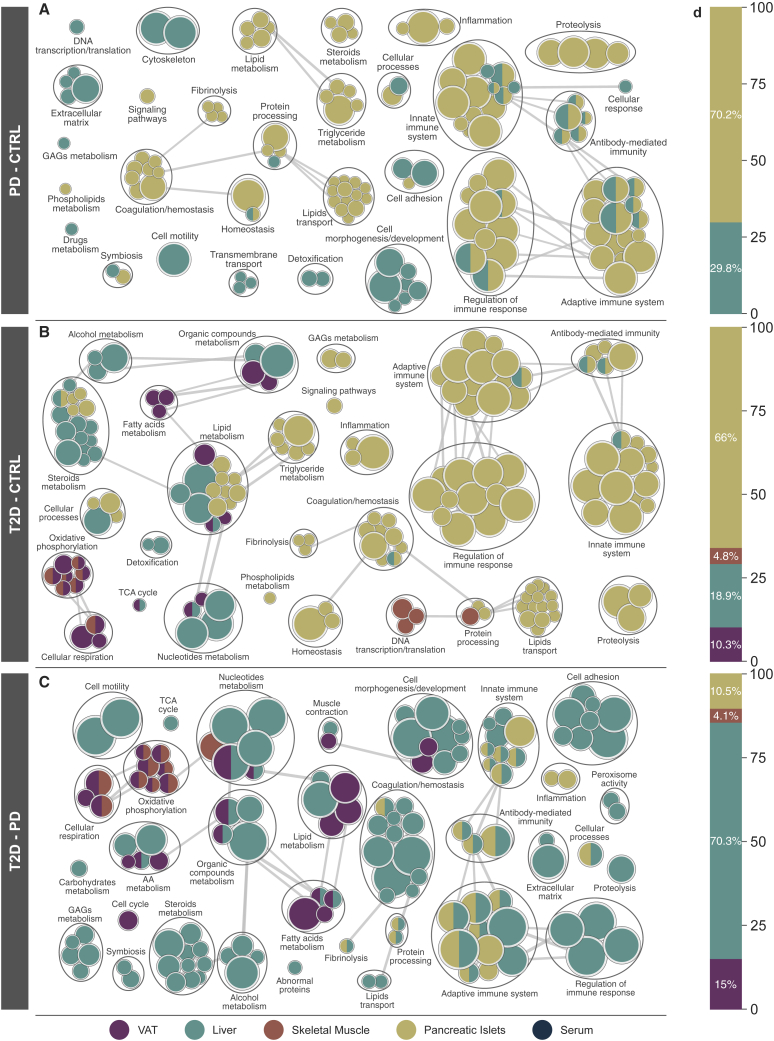
Figure 2.
Overview of enriched GO biological processes across tissues for pairwise comparisons among CTRL, PD, and T2D
(A–C) Networks were created using the Cytoscape plugins EnrichmentMap and AutoAnnotate based on a curated set of custom classes for GO biological processes (Table S1M).28, 29, 30 Nodes represent enriched GO biological processes (q < 0.01), and their size indicates the number of proteins in the biological process. Thickness of edges represents the fraction of shared proteins between nodes. Only edges describing high similarity were retained. Groups of nodes represent broader categories of individual GO terms. Panels show comparisons of (A) PD-CTRL, (B) T2D-CTRL, and (C) T2D-PD.
(D) Distribution of the enriched GO biological processes across tissues corresponding to the networks on the left-hand side. The distribution is presented as the fraction of enriched terms with respect to the total number of enriched terms for the pairwise comparison.