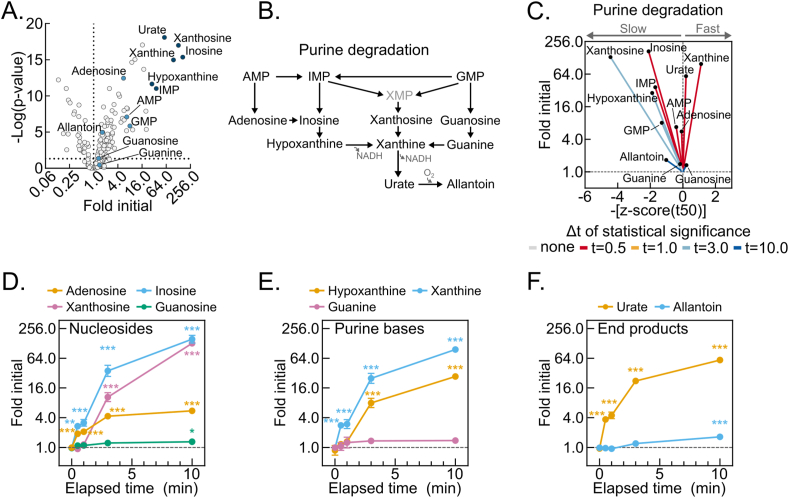
Figure 4.
Post-dissection liver hypoxia leads to enormous increases in purine degradation metabolites. (A) Volcano plot showing the interaction between metabolite fold changes and associated p-values at 10-minutes delayed freezing compared to time zero. Purine degradation pathway metabolite data points are labeled and colored blue, with the six most changed colored dark blue. The -log transformed p-values were calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 5–6 biological replicates). (B) Schematic of the purine degradation pathway. (C) Urchin plots for the purine degradation pathway showing line slopes as an interaction between the t50 kinetics and dynamic ranges of metabolite fold changes with delayed freezing. Metabolites were converted to line vectors displaying dynamic range (10-minute fold change compared to time zero; y-axis) and the negative z-score of the time required for one-half maximum fold change (-[z-score (t50)]; x-axis). Line color indicates the time-point by which statistically significant change was reached calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 5–6 biological replicates). (D–F) Line graphs showing the purine nucleosides (D), purine bases (E), and purine degradation end-products (F) fold changes by delayed freezing time-point compared to time zero. Data are presented as the mean ± SEM. P-values were calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 6 biological replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Related to Tables S1 and S2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)