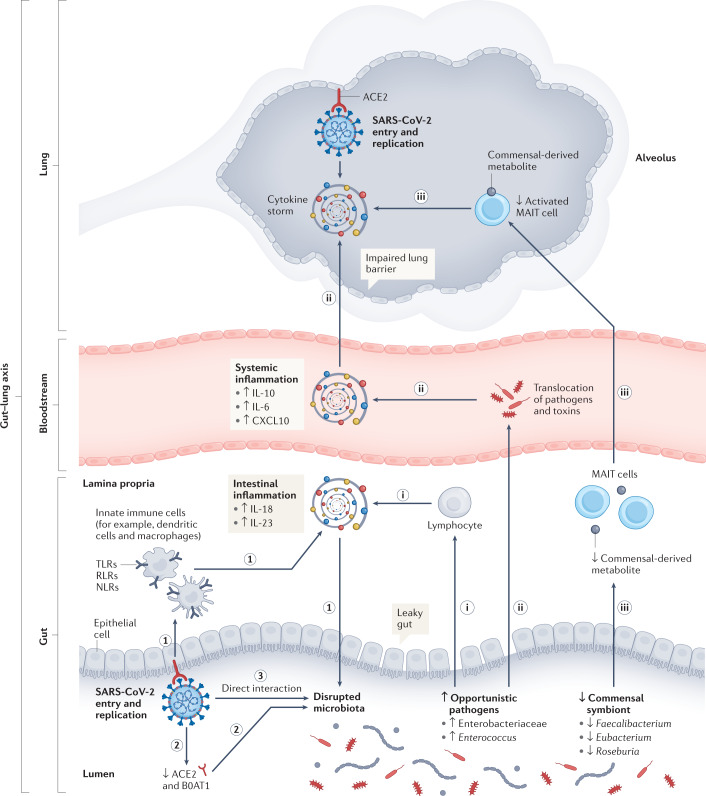
Fig. 1. Possible roles of the gut microbiota in dysfunctional immune responses and COVID-19 severity.
We propose potential mechanisms by which the gut microbiota can contribute to dysfunctional immune responses and coronavirus disease 2019 (COVID-19) severity. The first part hypothesizes that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection could lead to gut dysbiosis by several possible mechanisms: (1) invasion of SARS-CoV-2 can activate pattern-recognition receptors (Toll-like receptors (TLRs), RLRs, NLRs), which are recognized by innate immune cells, resulting in the release of various pro-inflammatory cytokines109. These activated immune responses could impair gut permeability, disrupt gut microbiota equilibrium and result in an increased abundance of opportunistic pathogens (for example, Enterobacteriaceae and Enterococcus) and decreased abundance of commensal symbionts (for example, Faecalibacterium, Eubacterium and Roseburia). (2) SARS-CoV-2 infection was found to downregulate the expression of angiotensin-converting enzyme 2 (ACE2) and B0AT1 (a molecular ACE2 chaperone) on the luminal surfaces of intestinal epithelial cells, which may facilitate the growth of the pathogens25,113,114. (3) An in vitro study found that SARS-CoV-2 might directly infect bacteria115. In the second part, we propose that specific intrinsic ‘microbiome signatures’ at the point of SARS-CoV-2 infection could influence the severity of infection and host immune response by several putative mechanisms: (i) increased opportunistic pathogens might be further recognized by innate lymphocytes and intensify gut pro-inflammatory responses185. (ii) Opportunistic pathogens and toxins could translocate into the circulatory system, causing bacteraemia and exacerbating systematic inflammation and disease severity20,21. (iii) Depleted commensal symbionts could negatively influence the recruitment of immune cells, such as activated mucosal-associated invariant T (MAIT) cells, to affect susceptibility and severity of respiratory tract infections22–24. CXCL10, C-X-C motif ligand 10.