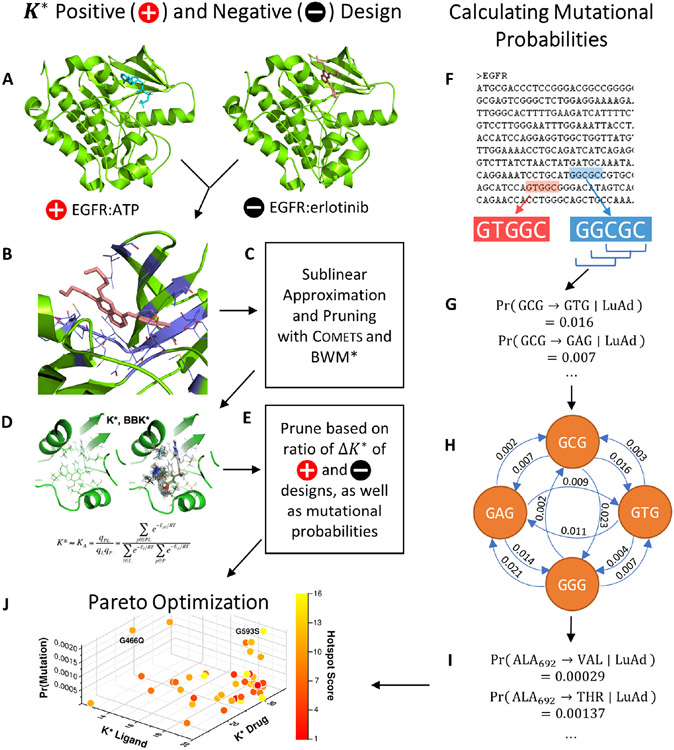
Figure 1. An example Resistor workflow with EGFR.
Resistor finds the Pareto frontier from OSPREY positive and negative designs, mutational probabilities, and resistance hotspots. (A) Two structures are required as input to OSPREY to compute postive and negative design K* scores. The structure for positive design is EGFR (green) bound to its endogenous ligand ATP (blue), for the negative design EGFR is bound to the drug erlotinib (pink). The goal of positive (resp. negative) design is to improve (resp. ablate) binding affinity. A mutation is resistant when its ratio of positive to negative K* scores increases. (B) All residues within 5 Å (purple) of the drug are allowed to mutate to any other amino acid. (C) Comets is used as an efficient, sublinear algorithm to quickly prune infeasible mutations. BWM* is used with a fixed branch width to compute a polynomial-time approximation to the K* score. (D) Candidate mutations that pass the Comets pruning step have their positive and negative K* scores computed in OSPREY. We recommend using the BBK* with MARK* algorithm as it is the fastest for computing K* scores. (E) Candidate resistance mutations are pruned when their ratio of positive to negative K* scores indicates a mutation does not cause resistance or if the target amino acid requires a mutation in all three DNA bases. (F) Resistor computes mutational probabilities using a protein’s coding DNA along with cancer-specific trinucleotide mutational probabilities for lung adenocarcinoma (abbreviated as LuAd), sliding a window (G) over 5′ - and 3′ -flanked codons. (H) Resistor employs a recursive graph algorithm to compute the probability that a particular amino acid will mutate to another amino acid (I). (J) Finally, Resistor uses Pareto optimization on the positive and negative K* scores, the mutational probabilities, and hotspot counts to predict resistance mutants.