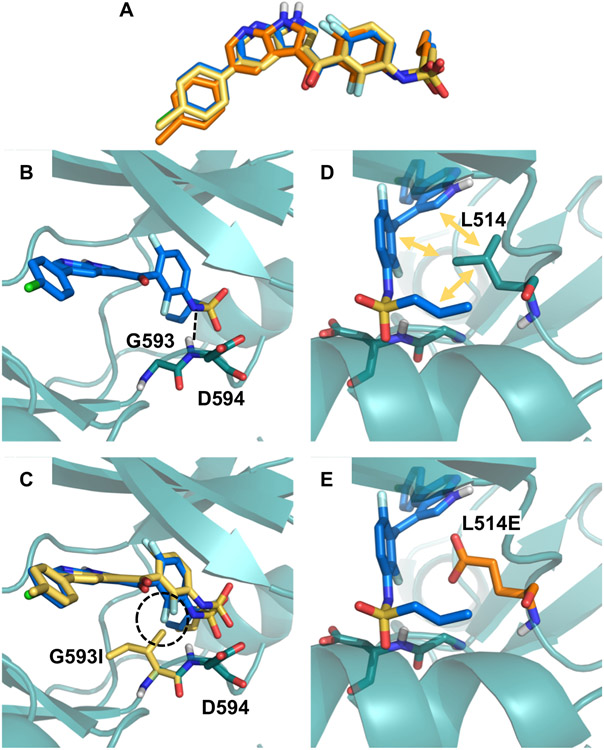
Figure 4. Structural analysis of BRAF mutations G593I and L514E.
(A) No major movements were required for vemurafenib to bind to the G593I (yellow) and L514E (orange) mutation in comparison to the wild type binding pose (blue). (B) BRAF G593 is located on the N-terminus of the activation loop and may facilitate conformational changes required to accommodate the vemurafenib propyl sulfonamide moiety in the back of the pocket. The backbone of the neighboring D594 residue interacts with the sulfonamide nitrogen of vemurafenib as indicated by black dashed lines. (C) Mutation of G593 to L not only restricts flexibility of the loop, but also puts the leucine side chain in too close proximity to the fluoro-substituted phenyl ring (highlighted with the dashed circle). (D) Residue L514 is involved in a variety of hydrophobic contacts with vemurafenib (indicated by yellow arrows), which are lost in the L514E mutant (E).