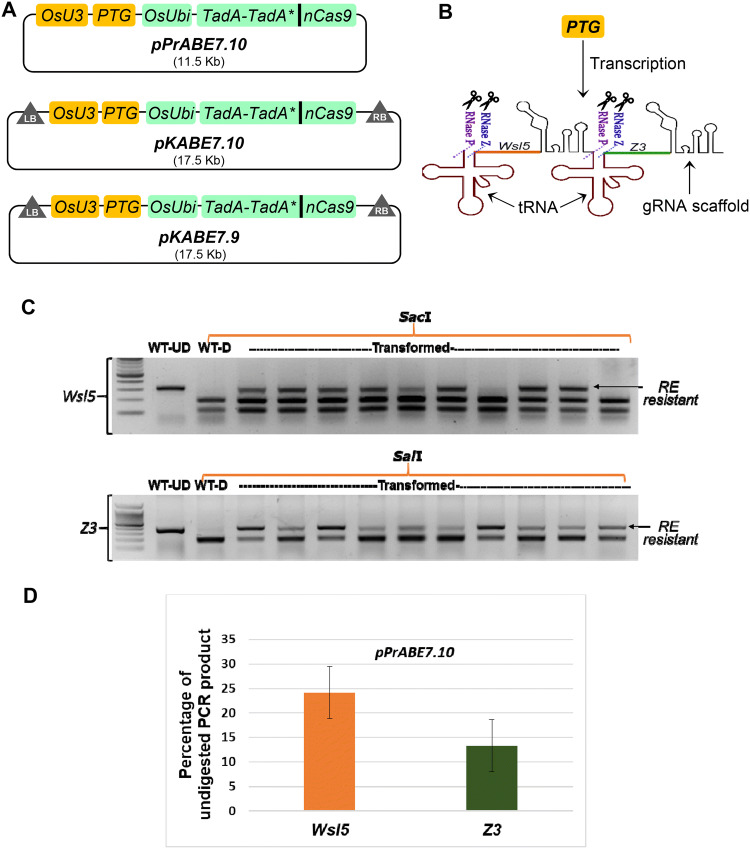
Fig. 1.
A → G base editing efficiencies at two target loci in transient assays using rice protoplasts. A Schematic diagram of plant adenine base editors constructed and used in this study. pPrABE7.10 was used in protoplast transformation; pKABE7.10 and pKABE7.9 were used for Agrobacterium-mediated calli transformation. B Diagrammatic representation of how PTG (polycistronic tRNA-gRNA) gene works in generating more than one single guide RNA (sgRNA). Endogenous RNase P and RNase Z splice out the tRNA releasing sgRNAs for targeting WSL5 (orange protospacer) and Z3 (green protospacer) loci. C Representative image of a 2% agarose gel showing a restriction enzyme (RE) digest of PCR amplicons spanning the target loci. RE resistant band indicates disruption of recognition sites by base editing. SacI and SalI enzymes were used for Wsl5 and Z3, respectively. WT-UD, wild type undigested/untreated; WT-D, wild type digested with RE. D Percentage of undigested PCR product after overnight incubation with RE. Bar diagram represents result for n = 15 ± SE