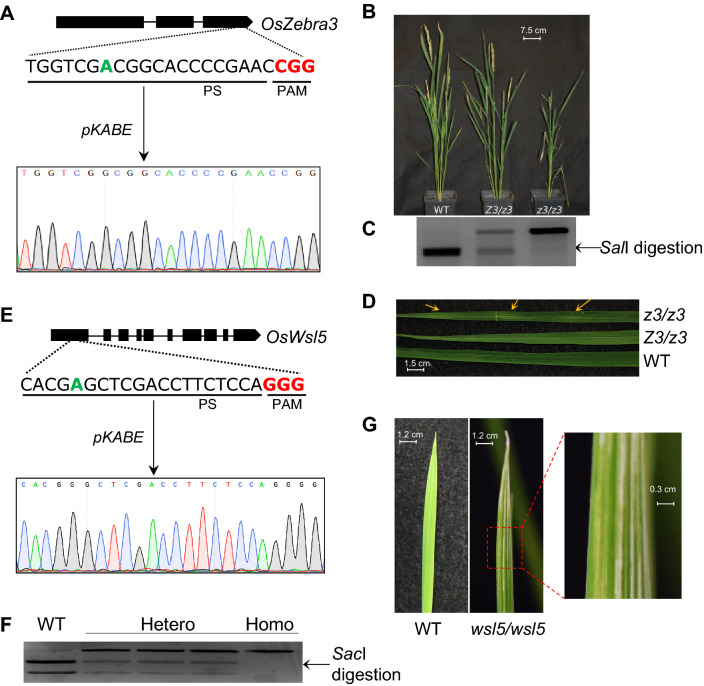
Fig. 3.
Characteristic phenotypes of z3 and wsl5 mutants generated by adenine base editors. A Schematic representation of the target site in OsZ3 locus and DNA chromatogram of homozygous mutant (z3/z3) plant. Green bold letter is the target base and red bold letters are the protospcaer adjacent motif (PAM). B Phenotypic appearance of wild type (WT), heterozygous (Z3/z3), and homozygous (z3/z3) mutant plants. C Representative restriction analysis from WT, Z3/z3, and z3/z3 plant showing complete, partial and nil digestion, respectively. D Leaf variegation phenotype showing green and dark green sectors in mature leaf. Yellow arrow indicates green sectors. E Schematic representation of the target site in OsWSL5 locus and DNA chromatogram of homozygous mutant (wsl5/wsl5) plant. Green bold letter is the target base and red bold letters are the protospcaer adjacent motif (PAM). F Representative RE analysis for WT, hetero-, and homozygous mutant plants. G White strip leaf phenotype in homozygous (wsl5/wsl5) seedling