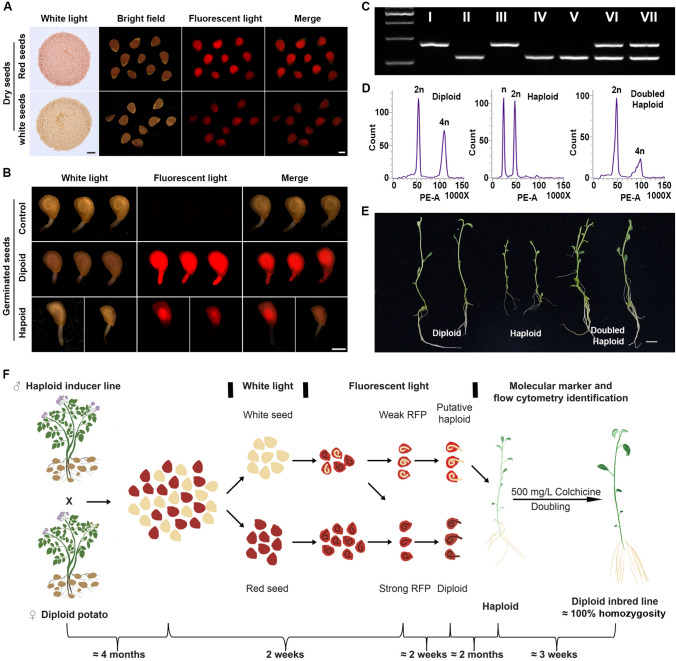
Fig. 1.
Construction of homozygous diploid potato via stdmp-based maternal haploid induction. A, B FAST-Red-based haploid seed identification. Under white light, the hybrid seeds could be divided into white seeds and red seeds, due to the presence of the FAST-Red marker. Some of the white seeds showed weak RFP signal under fluorescent light (A). During seed germination, putative haploids did not show RFP signal in the embryo root tip (B). C Seedlings from putative haploids were genotyped with a polymorphic marker between the inducer line and testers. The Roman numerals I to VII represent the bands from S15-65 (I), PG6359 (II), stdmp mutants (III), haploids from PG6359 × stdmp (IV and V), and diploids from PG6359 × stdmp (VI and VII). D Flow cytometry ploidy verification of the diploid control, putative haploid, and doubled haploid. E Phenotypes of the haploid, diploid, and doubled haploid. Haploid plants had smaller vegetative organs than diploid or doubled haploid plants. F Schematic overview of homozygous diploid potato generation, using the stdmp inducer line. Hybrid seeds could be easily divided into white and red seeds, under white light. Under fluorescent light, a few white seeds with weak RFP signal were selected for further analysis. Germinated seeds were checked for the absence/presence of RFP signal in the root tip. Seeds with no RFP signal in the emerging root tip are putative haploids and were further confirmed by molecular markers and ploidy analysis. True haploid plants were treated with colchicine to obtain doubled haploids. Scale bars: 1 cm (A, white light and E) and 1 mm (A, fluorescent light and B)