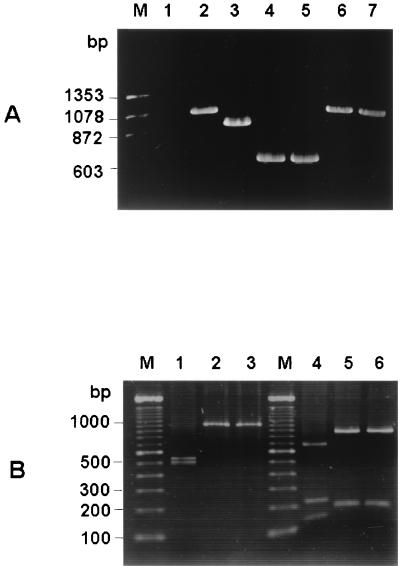
FIG. 4.
(A) Differential amplifications of the hypervariable region of rodent coronaviruses. Total RNA was extracted from virus-infected cells 24 h postinfection. RT-PCR was performed with the primer pair which was used for amplification of fragment a (1stf and 1str in Table 1), and the PCR products were electrophoresed on a 1% agarose gel. Lanes: M, molecular weight marker; 1, uninfected cells; 2, SDAV-infected cells; 3, MHV A59-infected cells; 4, MHV JHM-infected cells; 5, MHV 2-infected cells; 6, BCV-infected cells; 7, HCV OC43 infected cells. (B) Restriction patterns of PCR fragment a from SDAV (lanes 1 and 4), BCV (lanes 2 and 5), and HCV OC43 (lanes 3 and 6). Lanes 1, 2, and 3, digestion patterns obtained with restriction endonuclease KpnI; lanes 4, 5, and 6, digestion patterns obtained with restriction endonuclease MfeI; lane M, 100-bp ladder molecular weight marker. For the sizes of the digested fragments, see the text.