Figure 2 |. Hormonal crosstalk through transcriptional regulation.
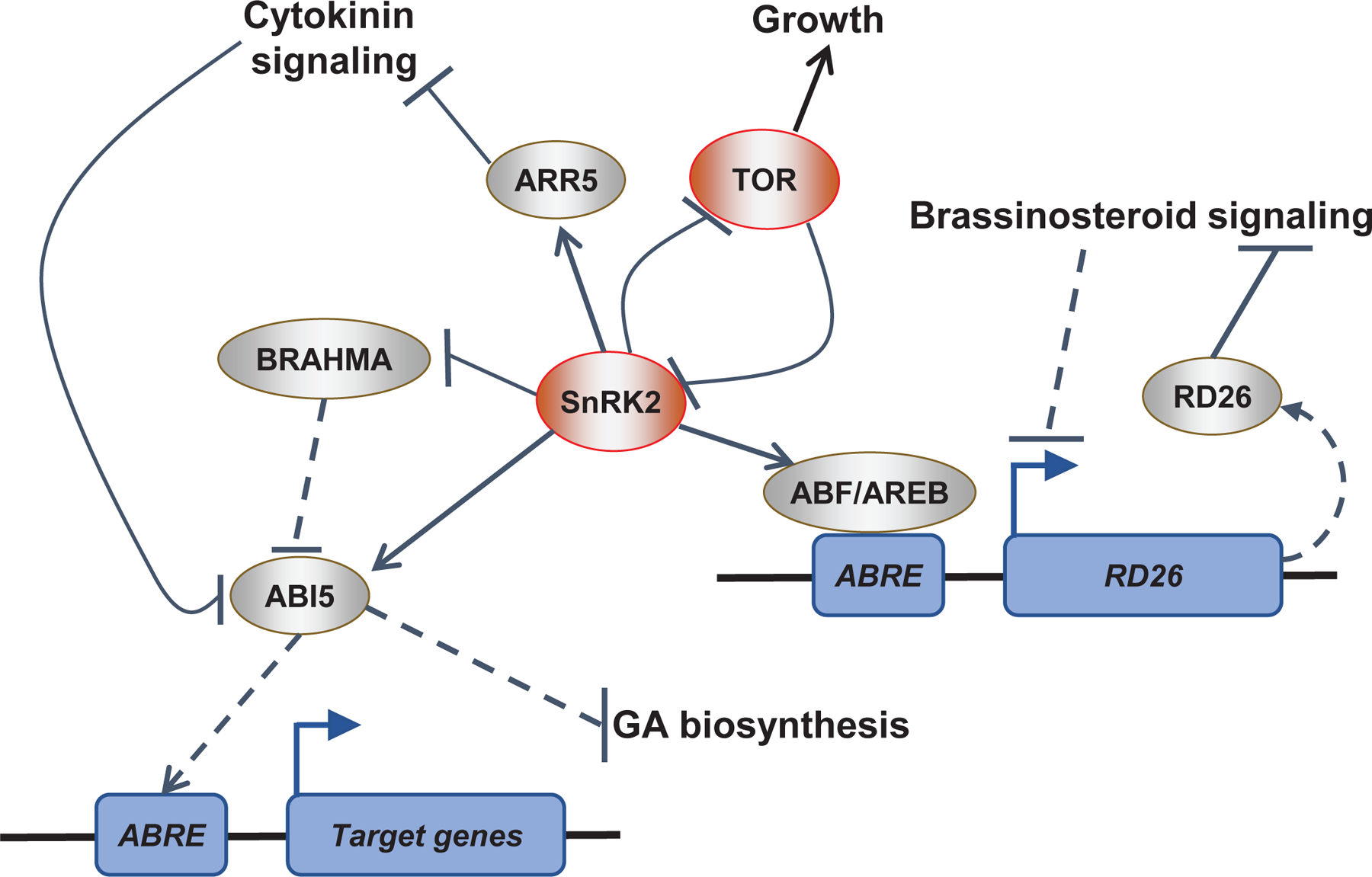
Plant hormones control abiotic stress response by altering transcriptional programs. The ABA signaling system intersects with many other hormone pathways during transcription. This figure summarizes the relationships between ABA signaling and key transcriptional regulators during hormonal signaling. During ABA-responses, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2 (SnRK2) protein kinases phosphorylate ABA-RESPONSIVE ELEMENT (ABRE)-binding proteins/ABRE-binding factors (AREBs/ABFs) and ABA-INSENSITIVE 5 (ABI5) transcription factors. AREBs/ABFs and ABI5 activate target genes with ABREs in their promotors to drive ABA responses. For instance, during drought stress AREBs/ABFs activate transcription of RESPONSE TO DESSICATION 26 (RD26), a transcription factor that can repress brassinosteroid signaling. Additionally, in dormant seeds, ABI5 target genes repress gibberellic acid (GA) biosynthesis and thereby block germination. Cytokinin signaling can repress ABA responses possibly by triggering the degradation of ABI5. ABA-activated SnRK2-type protein kinases promote transcriptional ABA responses by phosphorylating type-A ARABIDOPSIS RESPONSE REGULATOR5 (ARR5), a negative regulator of the cytokinin pathway. Finally, in unstressed conditions the protein kinase TARGET OF RAPAMYCIN (TOR) promotes plant growth by inhibiting SnRK2-type protein kinase-mediated ABA responses through phosphorylation of ABA receptors. Conversely, during stress SnRK2-type protein kinases phosphorylate and inhibit the TOR regulatory protein RAPTOR1B leading to growth repression. Note that not all mechanisms shown here are necessarily present at the same time or in the same cell/tissue.
