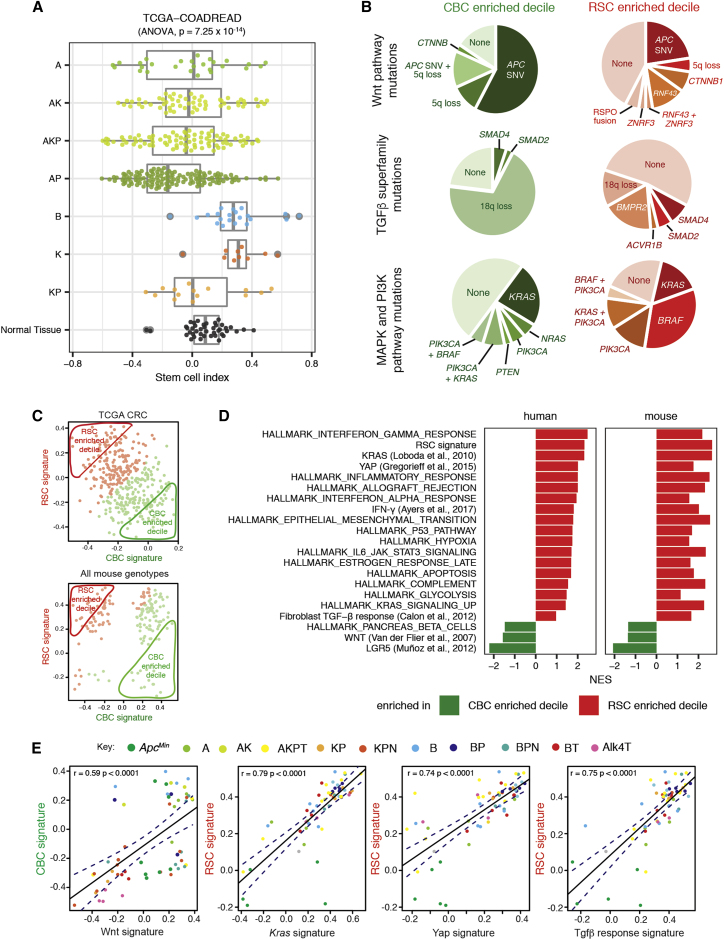
Figure 3.
Driver genes and pathways associated with variable stem cell molecular phenotype
(A) Human genotype-stem cell phenotype correlation based on stem cell index distribution in TCGA tumors with different putative driver gene single-nucleotide variant (SNV) mutation genotypes, contrasted to normal tissue from same dataset (driver gene initials: A is APC, K is KRAS, P is p53, B is BRAF).
(B) Comparison of mutation type and prevalence disrupting the Wnt pathway, MAPK and PIK3CA pathways, and the TGFβ superfamily in TCGA tumors subdivided into CBC- and RSC-predominant deciles.
(C) Segregation of mouse and human lesions by CBC (x axis) and RSC (y axis) signature expression. Predominant (above median) expression signature in each tumor is defined by color (CBC in green and RSC in red), and the 10% most polarized CBC- or RSC-expressing tumors were segregated into CBC- and RSC-enriched deciles for comparison.
(D) Gene set enrichment analysis of hallmark and select pathways in bulk transcriptome from human tumors (TCGA) and murine lesions (Glasgow dataset) segregated into CBC- and RSC-predominant deciles. Pathways shown have PFDR ≤ 0.25 apart from YAP in the mouse lesions and Fibroblast TGFβ response in the human tumors.
(E) Correlation of key pathway expression signatures with CBC or RSC gene expression across a range of mouse models. Different genotypes are identified by different colors as determined by the key. Driver alleles initialization: A is Apcfl/+, ApcMin is ApcMin, B is BrafV600E, K is KrasG12D, P is p53fl/fl, T is Tgfβr1fl/fl, N is Rosa26N1icd/+, Alk4 is Alk4fl/fl.