FIGURE 3.
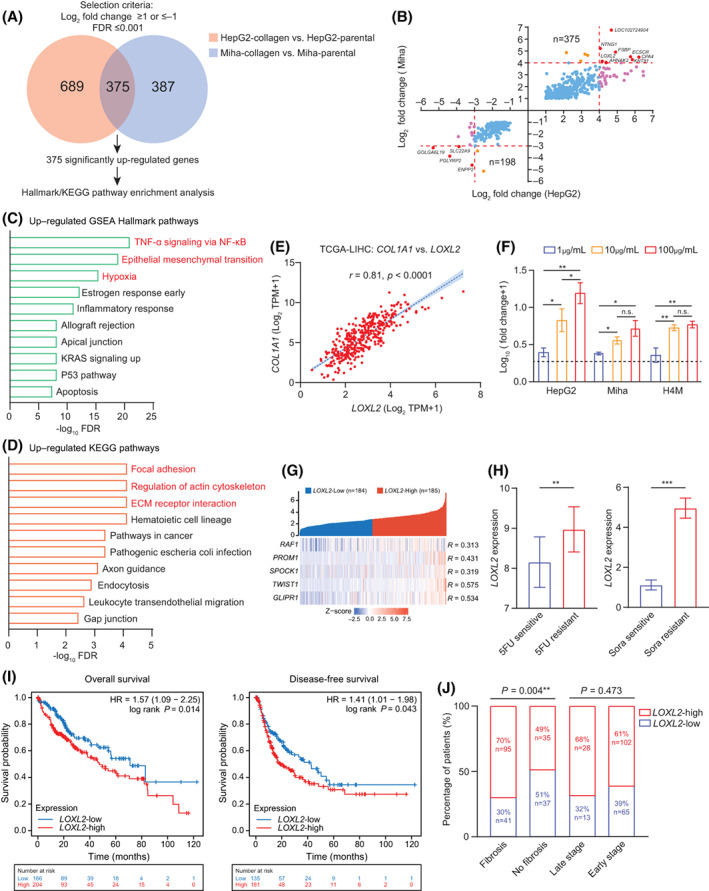
Collagen accumulation up‐regulated an ECM remodeler, LOXL2, in liver cancer cells. (A) Venn diagram shows 375 overlapped genes in 689 up‐regulated genes in collagen‐residing HepG2 cells and 387 up‐regulated genes in collagen‐residing Miha cells. (B) Transcriptome analysis identified 375 co‐up‐regulated and 198 co‐down‐regulated genes, including eight significantly co‐up‐regulated genes (LOXL2, AHNAK2, KRT81, CPA4, ECSCR, FSBP, NTNG1, and LOC102724904) and four significantly co‐down‐regulated genes (SLC22A9, PGLYRP2, ENPP2, and GOLGA6L19). (C) Top10 hallmark pathways enriched in collagen‐residing cells, ordered by –log10 FDR. (D) Top10 KEGG pathways enriched in collagen‐residing cells, ordered by –log10 FDR. (E) Pearson's correlation between LOXL2 and COL1A1 in TCGA‐LIHC (n = 369). (F) Fold change of LOXL2 expression in cells cultured on different densities of collagen, normalized by the LOXL2 expression of parental cells cultured on a noncollagen‐coated plate. (G) Pearson's correlation between LOXL2 and the five chemoresistant gene signature in TCGA‐LIHC (n = 369). (H) LOXL2 expression in patients treated with sorafenib (sensitive n = 21, resistant n = 46) and 5FU (sensitive n = 8, resistant n = 15) from GSE109211 and GSE28702. (I) Prognostic value of LOXL2 expression in overall survival and disease‐free survival of patients with liver cancer (n = 369). (J) Correlation between LOXL2 expression and liver fibrosis and the tumor stage. *p < 0.05, **p < 0.01, ***p < 0.001. AHNAK2, AHNAK nucleoprotein 2; COL1A1, collagen type I, alpha 1; ECM, extracellular matrix; CPA4, carboxypeptidase A4; ECSCR, endothelial cell surface expressed chemotaxis and apoptosis regulator; ENPP2, ectonucleotide pyrophosphatase/phosphodiesterase 2; FDR, false discovery rate; FSBP, fibrinogen silencer binding protein; GLIPR1, GLI pathogenesis related 1; GOLGA6L19, golgin A6 family like 19; GSE, gene expression data series; HR, hazard ratio; KEGG, Kyoto Encyclopedia of Genes and Genomes; KRAS, KRAS proto‐oncogene, guanosine triphosphatase; KRT81, keratin 81; LIHC, liver hepatocellular carcinoma; LOC102724904, methylenetetrahydrofolate dehydrogenase (NADP+ Dependent) 1 like pseudogene; LOXL2, lysyl oxidase‐like 2; NF‐κB, nuclear factor kappa B; n.s., not significant; NTNG1, netrin G1; PGLYRP2, peptidoglycan recognition protein 2; PROM1, prominin 1; RAF1, raf‐1 proto‐oncogene, serine/threonine kinase; SLC22A9, solute carrier family 22 member 9; SPARC, secreted protein acidic and cysteine rich (also known as osteonectin); SPOCK1, cwcv‐ and kazal‐like domains proteoglycan 1; TCGA, The Cancer Genome Atlas; TNF, tumor necrosis factor; TPM, transcript per million; TWIST1, twist family bHLH transcription factor 1
