Abstract
HASPIN is a nuclear serine-threonine kinase originally identified in the mouse testis. Its 193 bp DNA promoter element (hereafter, 193PE) regulates bidirectional, synchronous gene expression in the germ cells of male mice. Recent studies have shown that Haspin is also expressed in trace amounts in somatic cells; HASPIN also functions in oocytes. Haspin expression is regulated by the tissue-specific methylation of Haspin genomic DNA regions, including somatic cells. This study investigated relationship between 193PE and DNA methylation by examining methylation status of transgenic mice carrying 193PE and a reporter gene. In somatic (liver) cells carrying the reporter gene, 193PE induced methylation as well as trace expression of the reporter gene. In the testis, 193PE induced hypomethylation and intense reporter gene expression. Expression of HASPIN in an egg was assessed using human chorionic gonadotrophin to induce ovulation in female transgenic mice. The results showed that 193PE induced tissue-specific methylation, which resulted in reporter gene expression in a mouse egg.
Keywords: Embryo, Haspin, Inner Cell Mass, Germ Cell, Oocyte
HASPIN is a nuclear serein/thyrosin (Ser/Thr) kinase, expression of which was originally identified in haploid germ cells (1). While intensely expressed in the testis, trace amounts have been detected in the other tissues (2). Because Haspin is conserved in many organisms, including plants and yeast, it is considered fundamental for cell survival (3, 4). Recently, in normal cells in vivo, its role in mitosis is functionally complemented by the other molecules, as no abnormality is observed in Haspindisrupted mice (5). It was also reported that HASPIN played an important role in the growth of cancer cells, due to the blockage of HASPIN effect by the corresponding inhibitors (6-8). Analysis of HASPIN expression in vivo is an important because HASPIN phosphorylated histone H3 at threonine 3 during chromosomal partitioning and was a target for tumor suppression. The mechanism of Haspin gene expression has previously been reported. Transgenic mice were generated with a DNA fragment linking the element located in193 bp upstream from the transcription start site to EGFP and Ds-Red, as a reporter gene. The reporter assay in mice showed that element located in the 193 bp DNA fragment regulated Haspin expression in the testis (9). A tissue-differentially methylated region (T-DMR) was also identified, located 600 bp upstream (from –641 to –517) of the transcriptional start site of Haspin (10). The T-DMR of Haspin was hypomethylated in male germ cells, but hypermethylated in somatic tissues. These observations indicated that the 193 bp DNA promoter element (193PE) induced tissue-specific Haspin expression, mediated by the tissue-specific methylation of the genomic region around Haspin. Here we showed that 193PE induced methylation of a reporter gene in the genomic DNA of somatic cells and testis hypomethylation, mimicking specific expression in male germ cells. It has also been shown to induce reporter gene expression in the egg and blastocyst inner cell mass.
A pBiPRO2 reporter gene previously constructed in our lab was used to investigate methylation status of transgenes located in genomic DNA in the liver and testis of mice (Fig .1) (9). The transgenic mice were bred under specific-pathogen-free conditions in the animal room at Nagasaki International University (Japan). The animals were euthanized by cervical dislocation just before the experiments. All animal experiments conformed to the Guide for the Care and Use of Laboratory Animals and they were approved by the Institutional Committee of Laboratory Animal Experimentation and Ethics of Nagasaki International University (128). Genomic DNA isolated from the testis and liver was purified by phenol extraction (1) and its methylation status was determined by bisulfite genomic sequencing (CHEMICON International, USA). Figure 2 shows the results of bisulfite genomic sequencing. It has been reported that the region around the endogenous Haspin genomic region is differentially methylated in tissues and the endogenous Haspin genomic region including T-DMR was hypomethylated in the testis (Upper bar in Fig.2) (10). The pBiPRO2 transgene was hypermethylated in the liver and hypomethylated in testis (lower part of Fig.2). These results indicated tissuespecific methylation of the genomic Haspin gene locus by the 193PE Haspin promoter element.
Fig 1.
Construction of the reporter plasmid BiPRO-2 for transgenic mice. The 193 bp DNA promoter element (193PE) was inserted into the EGFP and DsRed reporter genes. Arrows indicate directions of Haspin (white arrow), and mADE (black arrow) is a non-coding mRNA transcript (11, 12). The AseI-NheI DNA fragments containing the reporter genes were used as transgenes.
Fig 2.
Schematic presentation of the methylation status on the locus of endogenous Haspin and transgenes region of the reporter genes. The top box shows genomic region of endogenous Haspin and differentially methylated region (shading) (10). The bottom box shows transgene region of the reporter gene in the genome. The bars indicate methylation status on the reporter genes in the testis and liver. The vertical bars represent location of methylated CpG. Region around the endogenous Haspin genomic region was differentially methylated between tissues, consistent with the activity of 193PE. Arrows indicated transcription of each gene and ‘met’ and ‘stop’ are the translation initiation and stop codons, respectively. mADE is a non-coding mRNA (12).
Because HASPIN has also been reported to function in meiosis in mouse oocytes (13), we investigated whether 193PE also regulated its expression. Oocytes, arrested in the first meiotic prophase of female transgenic mice, were grown with gonadotropins and expression of the reporter gene in the ovaries was observed. Expression of the reporter gene EGPF was examined by Western blotting of mouse ovarian tissues from transgenic mice with ovarian hyperstimulation and non-hyperstimulation; detection was achieved using anti-GFP monoclonal antibody as described previously (9). Briefly, superovulation was induced in transgenic female mice by an intraperitoneal injection of 5 IU of pregnant mare serum gonadotropin, followed 46-48 hours later by 5 IU of human chorionic gonadotropin. The mice were euthanized 14-16 hours later and their ovaries were collected. Gene expression was analyzed by Western blotting after sonicating the tissues in TBS-T (100 mmol/L Tris-HCl [pH=7.5], 150 mmol/L NaCl and 0.1 w/v% Tween 20) buffer. An EGFP signal, and thus gene expression in response to 193PE activity, was observed only in the hyperstimulated ovaries (Fig .3), but not detected in ovaries without hyperstimulation (11). These results suggested that expression of the gene from 193PE was induced when oocyte division arrest was released with hyperstimulation.
Fig 3.
Analysis of EGFP reporter gene expression in the ovaries. EGPF expression was examined by Western blotting of mouse ovaries from transgenic mice with ovarian hyperstimulation and non-hyperstimulation. Signal detection was achieved using anti-EGFP monoclonal antibody. Ov and Ov HCG indicate non-hyperstimulated or hyperstimulated ovaries. The arrow indicates Mr 31k. The arrowhead indicates the position of the EGFP signals.
Next, cumulus-oocyte complexes collected from the ampullae of the uterine tubes were fertilized in vitro with wild type sperm in HTF medium. After 24 hours, the embryos were cultured in potassium simplex optimization medium (KSOM) and Haspin expression over the time was assessed by fluorescence microscope (Fig .4). In these post-ovulatory oocytes, 193PE induced expression of reporter genes and participated in postfertilization embryonic development, although it was unclear whether the results were attributable to ovulation or to transcribed maternal mRNA. EGFP was expressed in morula cells, but thereafter a signal was observed only in the inner cell mass (ICM), not in trophectoderm cells. Although transcription and translation were maintained in the oocytes for several days during embryogenesis (14), these observations suggested that Haspin gene expression was specifically maintained in ICM of the blastocyst. HASPIN may be expressed and play a role in proliferating cells. Reason of no expression of HASPIN in trophoblasts is unclear, but it is linked to the epigenetic changes and expression during the process of trophoblast and ICM differentiation.
Fig 4.
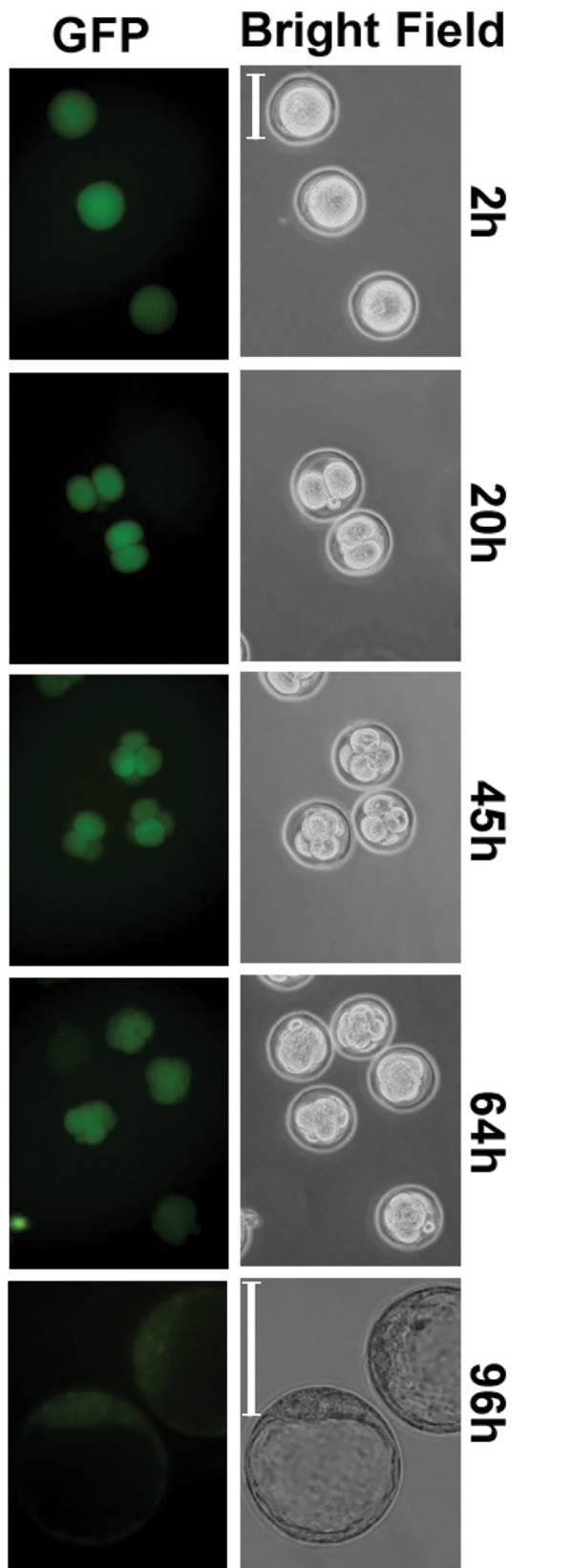
Fluorescence microscope images of the fertilized eggs of transgenic mice. The images were obtained at different times after in vitro insemination of the eggs. Green color indicates the EGFP expression. 2, 20, 45, 64 and 96 hours indicate time of the culture after insemination (scale bar: 100 mm).
In summary, 193PE from genomic Haspin induced tissue-specific methylation and specific gene expression in the surrounding genomic DNA region. In isolated mouse eggs, 193PE induced specific expression in the ICM of the blastocyst, confirming a role for this very small DNA fragment in tissue-specific methylation and gene expression in cells, other than germ cells of the testis. Further studies of the role of HASPIN and mechanism of tissue-specific methylation are needed. Use of transgenic mice carrying the EGFP reporter gene will aid in these investigations (15).
Acknowledgements
None of the authors have competing financial interests to declare. There was no financial support for this study.
Authors’ Contributions
H.T.; Contributed to the conception and design, experimental work, data and statistical analysis, data interpretation, and wrote the manuscript. K.T.; Contributed to experimental work, data analysis. Both authors read and approved the final version of the manuscript.
References
- 1.Tanaka H, Yoshimura Y, Nozaki M, Yomogida K, Tsuchida J, Tosaka Y, et al. Identification and characterization of a haploid germ cell-specific nuclear protein kinase (Haspin) in spermatid nuclei and its effects on somatic cells. J Biol Chem. 1999;274(24):17049–17057. doi: 10.1074/jbc.274.24.17049. [DOI] [PubMed] [Google Scholar]
- 2.Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19(4):472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins JM. Haspin-like proteins: a new family of evolutionarily conserved putative eukaryotic protein kinases. Protein Sci. 2001;10(8):167716–167784. doi: 10.1110/ps.49901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurihara D, Matsunaga S, Omura T, Higashiyama T, Fukui K. Identification and characterization of plant Haspin kinase as a histone H3 threonine kinase. BMC Plant Biol. 2011;11:73–73. doi: 10.1186/1471-2229-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada M, Goshima T, Matsuo H, Johmura Y, Haruta M, Murata K, et al. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat Commun. 2016;7:12059–12059. doi: 10.1038/ncomms12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huertas D, Soler M, Moreto J, Villanueva A, Martinez A, Vidal A, et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene. 2012;31(11):1408–1418. doi: 10.1038/onc.2011.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka H, Wada M, Park J. HASPIN kinase inhibitor CHR-6494 suppresses intestinal polyp development, cachexia, and hypogonadism in Apcmin/+ mice. Eur J Cancer Prev. 2020;29(6):481–485. doi: 10.1097/CEJ.0000000000000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elie J, Feizbakhsh O, Desban N, Josselin B, Baratte B, Bescond A, et al. Design of new disubstituted imidazo[1,2-b]pyridazine derivatives as selective Haspin inhibitors.Synthesis, binding mode and anticancer biological evaluation. J Enzyme Inhib Med Chem. 2020;35(1):1840–1853. doi: 10.1080/14756366.2020.1825408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tokuhiro K, Miyagawa Y, Yamada S, Hirose M, Ohta H, Nishimune Y, et al. The 193-base pair Gsg2 (haspin) promoter region regulates germ cell-specific expression bidirectionally and synchronously. Biol Reprod. 2007;76(3):407–414. doi: 10.1095/biolreprod.106.055236. [DOI] [PubMed] [Google Scholar]
- 10.Sato S, Maeda C, Hattori N, Yagi S, Tanaka S, Shiota K. DNA methylation-dependent modulator of Gsg2/Haspin gene expression. J Reprod Dev. 2011;57(4):526–533. doi: 10.1262/jrd.11-031a. [DOI] [PubMed] [Google Scholar]
- 11.Higgins JM. Structure, function and evolution of haspin and haspin-related proteins, a distinctive group of eukaryotic protein kinases. Cell Mol Life Sci. 2003;60(3):446–462. doi: 10.1007/s000180300038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins JM. The Haspin gene: location in an intron of the integrin alphaE gene, associated transcription of an integrin alphaEderived RNA and expression in diploid as well as haploid cells. Gene. 2001;267(1):55–69. doi: 10.1016/s0378-1119(01)00387-0. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen AL, Gentilello AS, Balboula AZ, Shrivastava V, Ohring J, Schindler K. Phosphorylation of threonine 3 on histone H3 by haspin kinase is required for meiosis I in mouse oocytes. J Cell Sci. 2014;127(23):5066–5078. doi: 10.1242/jcs.158840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winata CL, Korzh V. The translational regulation of maternal mRNAs in time and space. FEBS Lett. 2018;592(17):3007–3023. doi: 10.1002/1873-3468.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gohbara A, Katagiri K, Sato T, Kubota Y, Kagechika H, Araki Y, et al. In vitro murine spermatogenesis in an organ culture system. Biol Reprod. 2010;83(2):261–267. doi: 10.1095/biolreprod.110.083899. [DOI] [PubMed] [Google Scholar]





