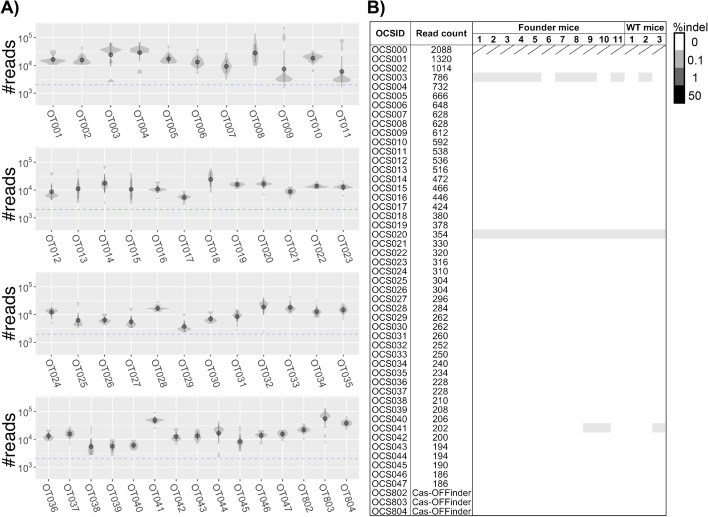
Fig. 3.
Validation of in vivo OT indels using targeted deep amplicon sequencing. A Targeted deep amplicon sequencing for OCSs. Violin plots show the distribution of the number of reads for each OCS (top 47 OCSs detected by CRISPR-KRISPR (OCS001~OCS0047) and three uniquely predicted sites by Cas-OFFinder (OCS802~OCS804)). Median distributions are shown by dark grey dots and lines, respectively. The dashed line indicates required minimum number of reads (2,000 reads). B Validation of 50 OCSs in 11 founder mice and three wildtype (WT) mice