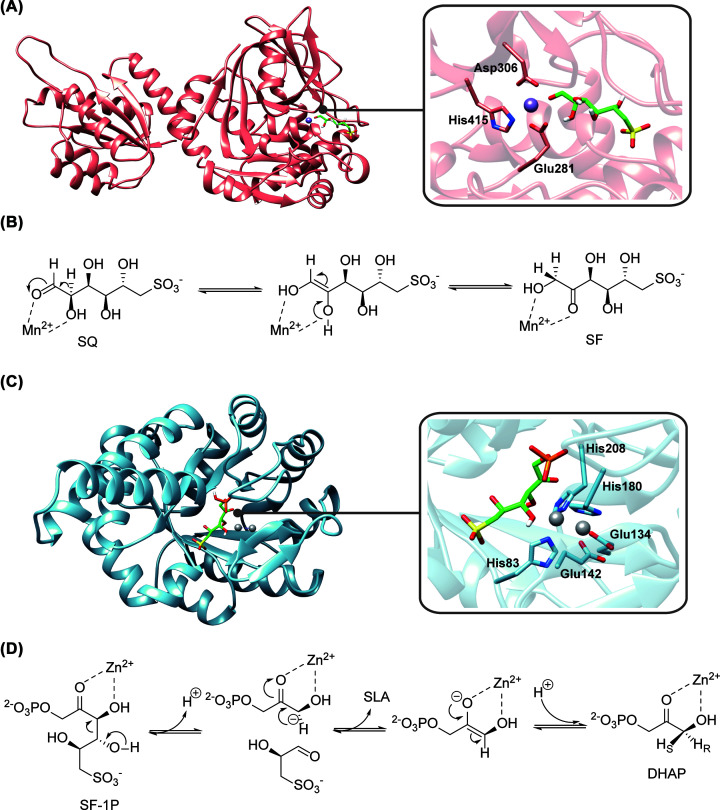
Figure 4. Structural models and proposed catalytic schemes for two key enzymes in the sulfo-EMP2 pathway.
(A) Structural model of SqvD (left) and the zoom-in view of the active site (right), showing conserved Mn2+-binding residues. (B) Proposed catalytic scheme for SqvD involving an ene-diol intermediate. (C) Structural model of SqiA (left) and the zoom-in view of the active site (right), showing conserved Zn2+-binding residues. The mobile Zn2+ cofactor occupies two mutually exclusive sites. (D) Proposed catalytic scheme for SqiA. Both models were constructed using AlphaFold2 [29] in ColabFold [30]. The putative Mn2+ site of SqvD and two putative Zn2+ sites of SqiA were estimated by alignment with the crystal structures of E. coli FucI (PDB: 1FUI) [28] and Helicobacter pylori class II fructose-1,6-biphosphate adolase (5UCK) [33], respectively.