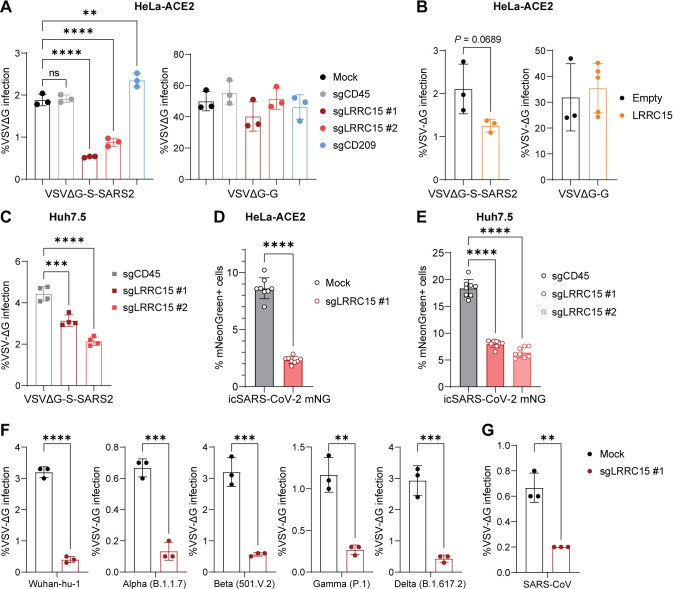
Fig 3. LRRC15 inhibits ACE2-mediated SARS-CoV-2 entry.
(A) HeLa-ACE2 cells were transduced with indicated activating sgRNAs and infected with VSV pseudoviruses, VSVΔG-S-SARS2 or VSVΔG-G. GFP signal was measured at 20 hpi by flow cytometry (n = 3). Representative of 3 independent experiments are shown. (B) HeLa-ACE2 cells expressing LRRC15 or empty vector were infected with VSV psuedoviruses and GFP signal was measured at 20 hpi by flow cytometry (n = 3). Representative of 3 independent experiments are shown. (C) Huh7.5 cells were transduced with indicated activating sgRNAs and infected with VSVΔG-S-SARS2. GFP signal was measured at 20 hpi by flow cytometry (n = 4). Representative of 3 independent experiments are shown. (D, E) HeLa-ACE2 (D) or Huh7.5 (E) were transduced with indicated activating sgRNAs and infected with icSARS-CoV-2 mNG at 1 MOI. Infected cell frequencies were measured by mNeonGreen expression at 24 hpi (n = 8). Representative of 2 (D) or 3 (E) independent experiments are shown. (F) LRRC15-induced or mock HeLa-ACE2 cells were infected with VSV pseudoviruses harboring spike proteins of different SARS-CoV-2 variants. GFP signal was measured at 20 hpi by flow cytometry (n = 3). (G) LRRC15-induced or mock HeLa-ACE2 cells were infected with VSV pseudoviruses harboring SARS-CoV-1 spike. GFP signal was measured at 20 hpi by flow cytometry (n = 3). Data represent means ± SD (A-G). Data were analyzed by one-way ANOVA with Dunnett multiple comparisons test (A, C, E) or unpaired two-tailed t test (B, D, F, G). ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. For underlying data, see S3 Data. ACE2, angiotensin-converting enzyme 2; GFP, green fluorescent protein; hpi, hours postinfection; LRRC15, leucin-rich repeat-containing 15; MOI, multiplicity of infection; SARS-CoV-1, Severe Acute Respiratory Syndrome Coronavirus; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; sgRNA, single guide RNA; VSV, vesicular stomatitis virus.