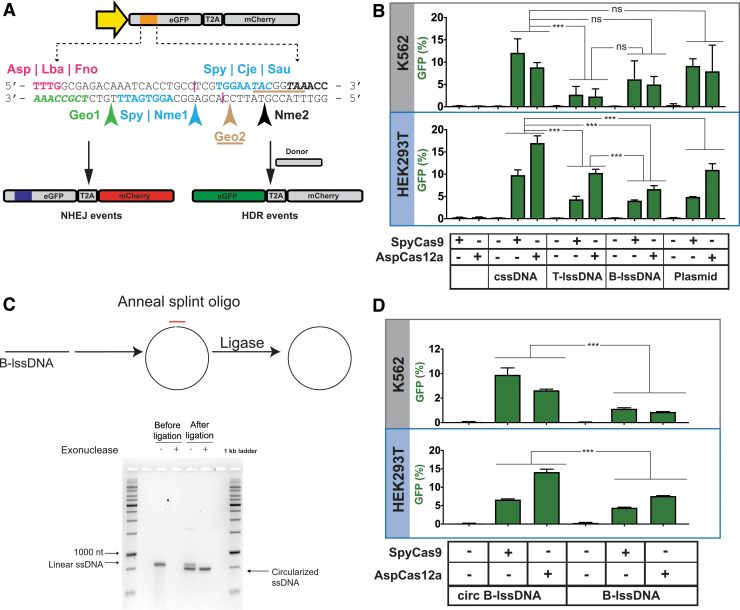
FIG. 1.
Comparisons of the integration efficiencies of different donor topologies on HDR using the TLR-MCV1 cassette in human cells. (A) The schematic depicts the TLR-MCV1 system showing the SFFV promoter driving the expression of GFP and mCherry, separated by a ribosome-skipping T2A signal. The yellow arrow depicts the SFFV promoter driving the expression of the GFP-T2A-mCherry cassette. The orange box indicates the insertion disrupting GFP containing target sequences for different Cas effectors. The sequence of insertion is shown below the schematic of TLR-MCV1. Sequences and arrows in blue indicate overlapping PAMs and a common cut site associated with SpyCas9, Nme1Cas9, CjeCas9, and SauCas9. The bolded black sequence and black arrow depict the Nme2Cas9 PAM and cut site, respectively. The magenta text shows PAMs associated with Cas12a effectors, and their approximate cut sites are shown by magenta lines. The PAMs associated with Geo1Cas9 and Geo2Cas9 are highlighted in green italicized text and brown-underlined italicized text, respectively. The cut sites for these two Cas9s are shown by green and brown arrows, respectively. DSBs at any of the sites may be imprecisely repaired through the NHEJ pathway resulting in mCherry expression (shown on the left) if repair results in productive translation due to a +1 frameshift. In the presence of donor DNA, HDR-mediated correction of the “broken” GFP region results in restoration of GFP expression (shown on the right). (B) Efficacy of distinct DNA templates in driving HDR. The graph depicts the percentage of GFP-positive cells obtained after codelivery of SpyCas9 or AspCas12a RNP with cssDNA, T-lssDNA, B-lssDNA, or plasmid DNA repair templates into TLR-MCV1 K562 cells (upper gray box) and TLR-MCV1 HEK293T cells (lower blue box). Bars represent the mean from three independent biological replicates and error bars represent the s.e.m. (C) Schematic of the approach used to generate circularized B-lssDNA. A short oligonucleotide (red) is hybridized to the B-lssDNA containing a 5′-phosphorylated end such that the oligo spans the 5′ and 3′ ends of the lssDNA. The sample is treated with Escherichia coli DNA ligase to ligate the ends. The lssDNA sample is then treated with exonucleases (I and III) to eliminate residual uncircularized lssDNA. The native agarose gel shows linear and ligated lssDNA before and after treatment with exonucleases, which digest unprotected, linear DNA species. (D) The graphs depict the percentage of GFP-positive cells obtained after codelivery of SpyCas9 with B-lssDNA and circularized B-lssDNA DNA repair templates into TLR-MCV1 K562 cells (upper gray box) and TLR-MCV1 HEK293T cells (lower blue box). Bars represent the mean from three independent biological replicates and error bars represent (s.e.m.). n.s., p value not significant; ***p < 0.001. B-lssDNA, biotin-based affinity purified linear ssDNA; cssDNA, circular ssDNA; DSB, double-strand break; GFP, green fluorescent protein; HDR, homology-directed repair; lssDNA, linear ssDNA; NHEJ, nonhomologous end joining; PAM, protospacer adjacent motif; RNP, ribonucleoprotein; s.e.m., standard error of the mean; ssDNA, single-stranded DNA; TLR-MCV1, traffic light reporter multi-Cas variant 1; T-lssDNA, reverse-transcription generated linear ssDNA.