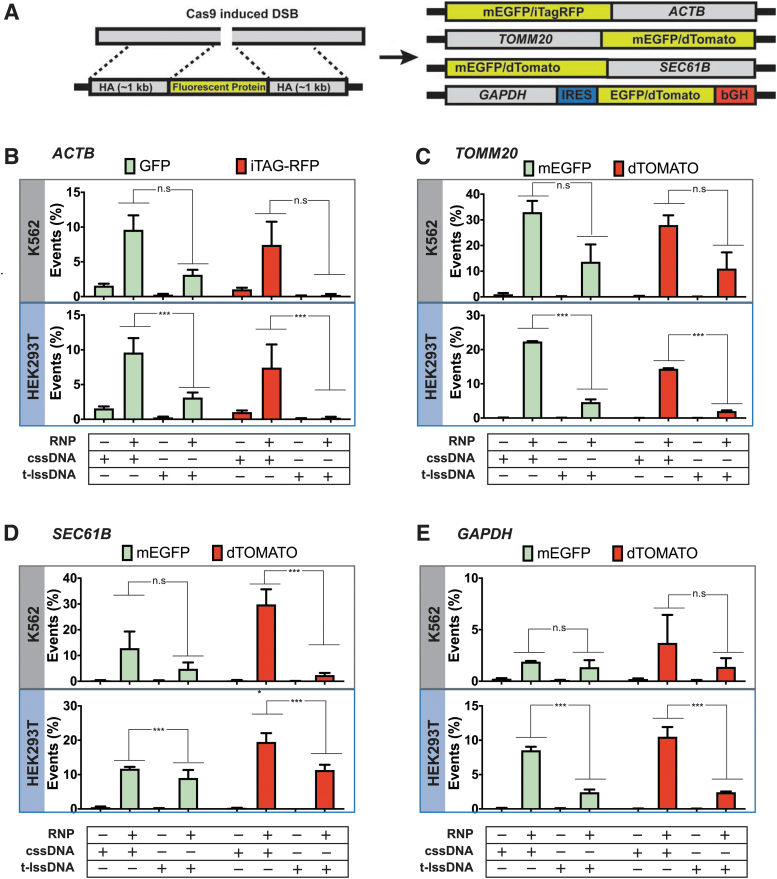
FIG. 3.
Comparison of the type of DNA donor on the efficiency of HDR at endogenous genomic loci in human cells. (A) Schematic of fluorescent protein tagging. The left panel shows a schematic of a genomic region containing the SpyCas9 target site and the design of a donor template containing the fluorescent protein of interest flanked by HA. The right panel shows a schematic of each target genomic locus and the arrangement of the fluorescent tag (EGFP, dTomato, or iTag-RFP) following integration. Three of the donors (targeting ACTB, TOMM20, and SEC61B) produce direct fusions of the tag to the endogenous protein. The donor designed to fluorescently tag the GAPDH locus contains an IRES and a bGH polyadenylation sequence. (B–E) Bar graphs displaying the percentages of fluorescent cells obtained upon codelivery of 20 pmoles of SpyCas9 complexed with 25 pmoles of guide-RNA targeting the (B) ACTB, (C) TOMM20, (D) SEC61B, or (E) GAPDH locus with or without cssDNA or T-lssDNA as a donor template. Bars represent the mean from three independent biological replicates and error bars represent s.e.m. n.s.: p value not significant; ***p < 0.001. bGH, bovine growth hormone; EGPF, enhanced green fluorescence protein; GAPDH, glyceraldehyde 3-phophate dehydrogenase; HA, homology arms; IRES, internal ribosome entry site; RFP, red fluorescent protein.