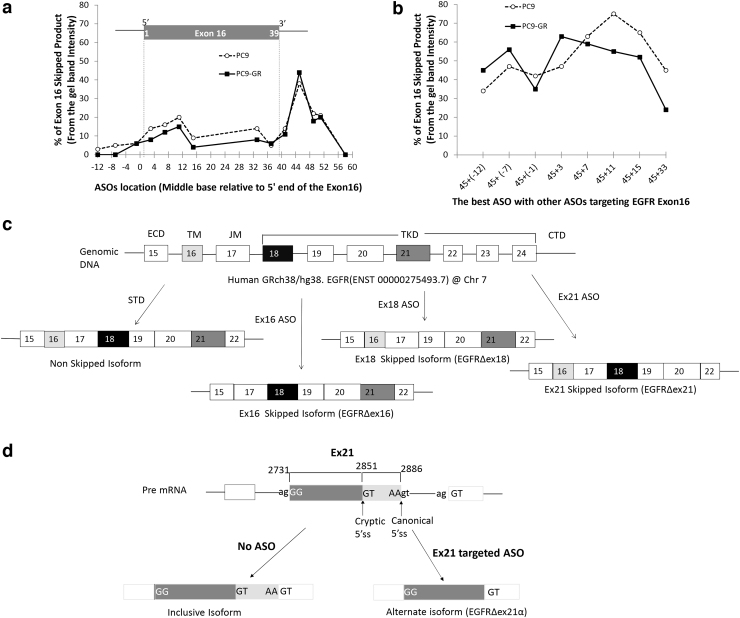
FIG. 1.
Optimization of ASO target sequence and schematic diagram of human EGFR with its expected splice variants. To optimize exon 16 skipping, we systematically designed and tested in vitro ASOs centered on every fourth base pair near the 5′ and 3′ ends of exon 16. The x-axis shows the base pair location of the center of each ASO target relative to the start of exon 16. PC9 (dashed line) and PC9-GR (solid line) cells were exposed to 5 μM of EGFR ASOs for 48 h and quantified with respect to percent EGFR exon 16 skipped product (skipped/total) relative to the wild-type isoform by RT-PCR. We first examined (a) single ASOs, which revealed a distinct peak in efficacy for both cell lines centered on exon base pair 45 located with the 5′ splice site of intron 16 and a secondary peak within the 5′ exon centered on base pair 11. We then examined whether the ASO centered on base pair 45 could be combined with a second ASO to provide a synergistic effect (b). Synergy was detected using a second ASO when it targeted the 5′ end of the exon (nonoverlapping) and a decreased efficacy when the second ASO was located near the original ASO (base pair 33) thought to be capable of sterically competing with the first ASO. (c) ASO targeting of exons 16 and 18 resulted in mRNA expression of the expected isoforms. In contrast, ASO targeting of exon 21 resulted in the use of a cryptic 5′ splice site formed within exon 21 that results in persistent inclusion of the 5′ portion of exon 21 (d). ASO, antisense oligonucleotide; EGFR, epidermal growth factor receptor; RT-PCR, reverse transcriptase–polymerase chain reaction.