Fig. 2. Non-RBD antibodies, selected to prioritize diversity, were used to identify non-RBD SARS-CoV-2 antibodies that bind SARS-CoV-1, a surrogate for epitope conservation.
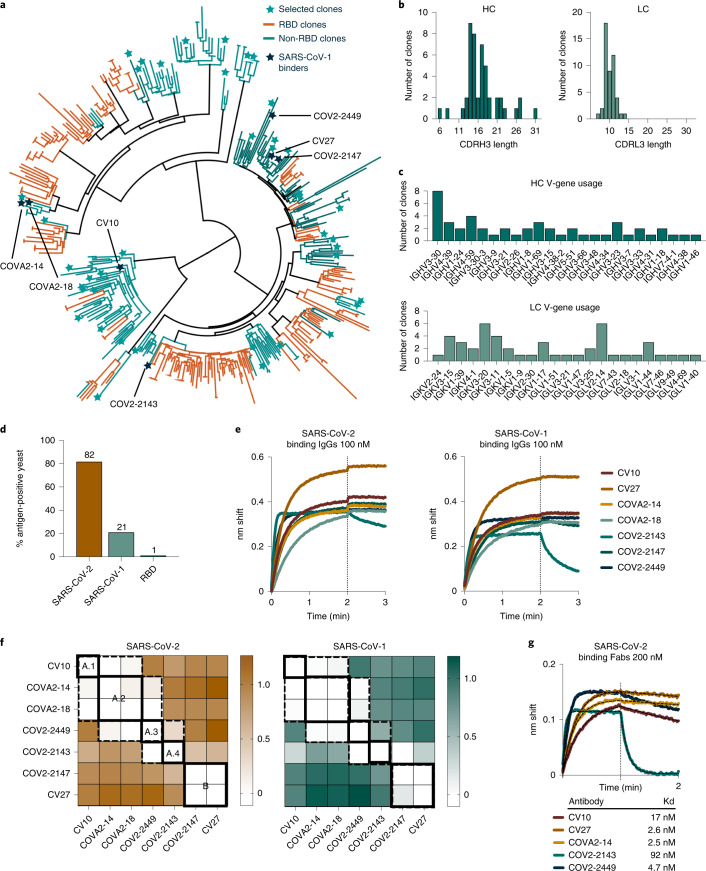
a, A phylogenetic tree of 422 HC sequences, from our curated library of 696 anti-SARS-COV-2 spike antibodies, generated using Geneious Prime. Germline alleles are not shown. Forty-eight selected clones are shown as stars. Histograms of the HC and LC (b) CDR3 lengths and (c) V-gene usage from the 48 selected non-RBD clones indicated in a. d, A binding profile of the scFv-yeast library produced from the sequences identified in a and Extended Data Fig. 1. e, BLI binding of identified cross-reactive clones expressed as IgGs at 100 nM to SARS-CoV-2 spike (left) or SARS-CoV-1 spike (right). f, BLI competition binding assay of the seven cross-reactive antibodies binding to SARS-CoV-2 (left) and SARS-CoV-1 (right). White indicates no binding of the tested antibody, indicating that the antibodies compete for binding. Antibodies that compete are surrounded by dotted lines; unique competition groups are surrounded by solid lines. The five unique competition groups are labeled on the SARS-CoV-2 binding competition map. Sites A.1–A.4 are indicated as an overlapping supersite. Loading antibodies are indicated in columns, and competing antibodies are indicated in rows. g, Binding of antibody Fab fragments at 200 nM against SARS-CoV-2 spike. Hashed lines show KD fit determined using Prism.