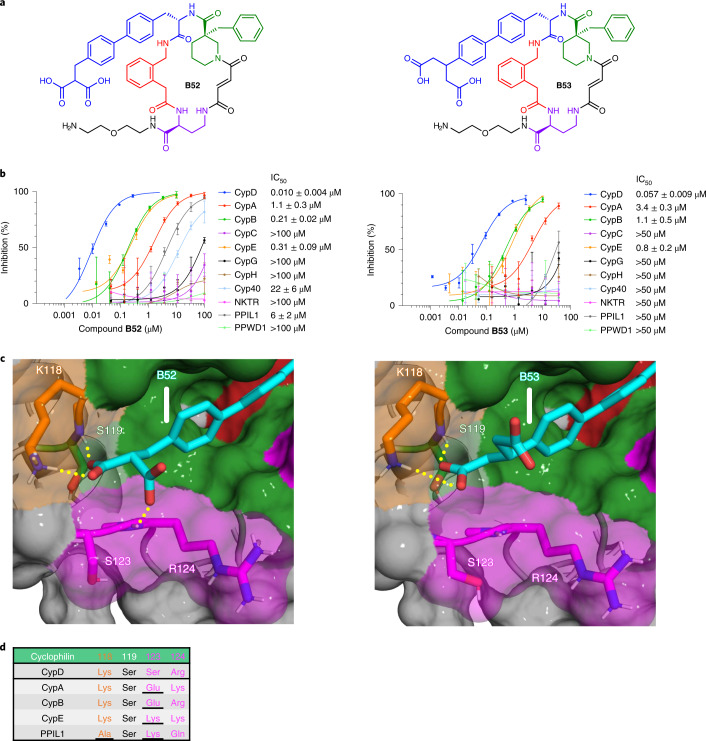
Fig. 4. Biphenyl dicarboxylates achieve strong CypD selectivity.
a, Structure of B52 and B53. b, Cyclophilin prolyl-isomerase inhibition screens for B52 and B53. c, Co-crystal structures of B52 (PDB ID 7THD, 1.16 Å resolution) and B53 (PDB ID 7THF, 1.10 Å resolution) bound to CypD, viewing the S2 pocket. Active site binding is identical to A26 (Fig. 1e). Yellow dashes indicate predicted hydrogen bonds. d, List of residues on the far side of the S2 pocket of cyclophilins that are proximal to the ligand carboxylates. Both compounds retain CypD potency similar to that of mono-carboxylate B23, while enhancing selectivity over CypA, CypB, CypE, and PPIL1. The malonic and glutaric acids of B52 and B53, respectively, position the carboxylate in a similar pose as B23 (Fig. 3c), while presenting a second carboxylate to the S123 residue. B52 forms a predicted hydrogen bond with the peptide backbone of S123–R124. R124 is pushed out of the S2 pocket, consistent with other macrocycles containing large S2-binding groups such as B1. B52 and B53 achieve selectivity over CypA and CypB through charge repulsion with a glutamate at the analogous 123 position, while creating a steric clash with PPIL1 and the lysine of CypE at this same position. IC50 values reflect mean ± s.d. of four independent replicates (each comprising three technical replicates). Graphs show a representative single independent replicate (independent replicate 1 is shown, containing three technical replicates) with data points and error bars reflecting mean ± s.d. of individual assays at one dose. Further independent replicates are shown in Supplementary Fig. 18b,c.